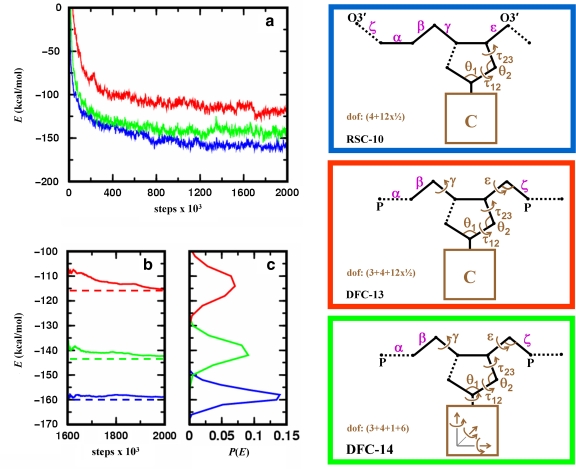
FIG. 6.
Analyzing energetic relaxation of chain breakage-closure Monte Carlo trajectories started from regular B-DNA conformation. Location of the chain break (dotted line) and the independent degrees of freedom, Xi (marked brown), for the three protocols used. In the first protocol (RSC-10, blue), MCRSC is employed on the closure problem of Figure 2b; the nucleotide conformation is changed by “natural” moves to give 10 independent degrees of freedom. In the second protocol (DFC-13, red), MCDFC is employed on P–O5′ bond closure; the nucleotide conformation is changed by “natural” moves with two additional main chain torsion angles and one additional main chain bond angle to give 13 independent degree of freedom. In the third protocol (DFC-14, green), MCDFC is employed on P–O5′ bond closure; the nucleotide conformations are changed by “physical” moves that include rigid body motion of the base to give 14 independent degrees of freedom. In all protocols, the dependent degrees of freedom, Xd, include torsion angle variables (magenta) that are depicted next to the bond around which the rotation is applied. Unless indicated otherwise, bond angles at all atoms along the chain also contribute to Xd. (a) The averaged value of the total energy as a function of Monte Carlo iterations where the averages are calculated from 10 independent runs. (b) The cumulative variation of the averaged total energy over the final 400,000 Monte Carlo iterations. (c) The distribution of the averaged total energy using the same data as in (b).