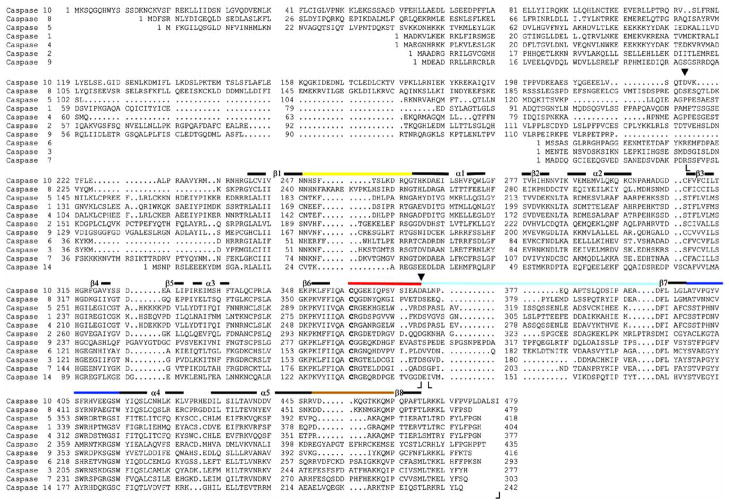
Fig. 2.
Sequence alignment of eleven human caspases. Amino acid residues in L1 (52–66) (yellow), L2 (163–175) (red), L3 (198–213) (blue), L4 (247–263) (brown) and L2′ (176′–192′) (cyan) of caspase–3 are shown by the solid lines, and the colors correspond to those in Figs. 5–8. The dark lines indicate regions of secondary structure. The catalytic cysteine (C163) is shown in bold. Boundaries observed in the crystal structures for the large subunit (S29-D175) and the small subunit (G177-H277) of caspase–3 are indicated by (⌊ ⌋ ). Two processing sites in procaspase–3 (D28 and D175) are indicated by (▾), where cleavage at D28 removes the prodomain and cleavage at D175 separates the two subunits, as described in the text. Sequence data were obtained using NCBI (http://www.ncbi.nlm.nih.gov/). Boundaries for the other caspases are approximately the same as those indicated for caspase-3. The sequence for caspase-10 is that of isoform 10/a.