Abstract
Background
The glycation of human serum albumin (HSA) during diabetes can affect the ability of this protein to bind drugs and small solutes in blood. This study describes the use of 16O/18O-labeling and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to compare the levels of modification that occur throughout HSA under various glycation conditions in vitro. These quantitative studies build on a recent report that has identified the early and advanced glycation products that are formed on such samples of HSA.
Methods
Glycated HSA samples were prepared by incubating 42 g/l HSA with 0 to 15 mmol/l glucose at pH 7.4 and 37°C for up to 5 weeks. A control HSA sample was digested in 16O-enriched water and glycated HSA samples were digested in the presence of 18O-enriched water. These 2 types of samples were then mixed and the amounts of 16O- vs. 18O-labeled peptides were measured to determine the levels of modification that were occurring throughout HSA.
Results
The largest levels of modification occurred in residues 101-119, 1-10 or 42-51, 87-100, 360-372, 521-531, and 275-286 of HSA after 2 weeks of glycation, and in residues 21-41, 1-10 or 42-51, 521-531, 82-93, and 146-160 after 5 weeks of glycation. Some of these regions contained the N-terminus, K199, K439, and K525, which have been previously identified as major glycation sites on HSA. The glycation pattern of HSA was dominated by early glycation products (e.g., fructosyl-lysine) after a reaction period of 2 weeks for mildly glycated HSA, while advanced glycation end products became more prominent at longer reaction times.
Conclusions
The time course of the observed modifications indicated that the pattern of glycation products changed as HSA was incubated over longer periods of time with glucose. Several regions found to have significant levels of modification were at or near the major drug binding regions on HSA. These results explain why the interaction of some drugs with HSA has been observed to vary with the level of glycation for this protein.
Keywords: Non-enzymatic glycation, Human serum albumin, Diabetes, Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, 16O/18O-Labeling, Quantitative proteomics
1. Introduction
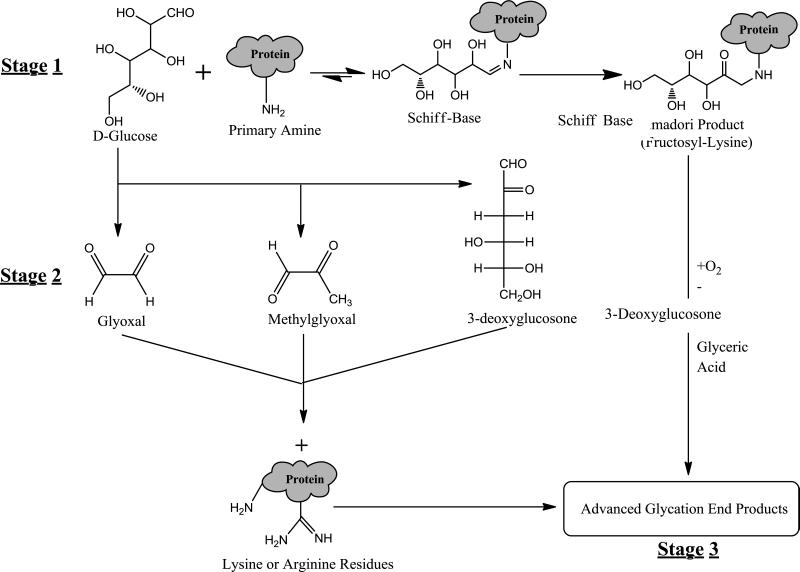
The non-enzymatic glycation of various proteins in the body has been linked to many of the chronic complications of diabetes, including nephropathy, macrovascular problems, and cataract formation [1-4]. These reactions and processes are typically accelerated during diabetes because of the increased blood sugar concentrations found in this disease [4,5]. The early stages of glycation involve the reaction of a reducing sugar, such as glucose, with primary amine groups on a protein [6-8]. This reaction leads to the formation of a Schiff base, which can later rearrange to form a more stable Amadori product (see Figure 1) [9,10]. Over time, reactive dicarbonyl compounds can form through the degradation of glucose or other free sugars (e.g., via retroaldol condensation and auto-oxidation) [11,12] and can combine with arginine or lysine residues on proteins to form advanced glycation end products (AGEs). Some AGEs can also form through the oxidation and rearrangement of Amadori products [13].
Figure 1.
Reaction scheme summarizing the various stages of glycation. The initial step (stage 1) is reductive amination, which results in the formation of a Schiff base followed by the creation of an Amadori product. The second stage is characterized by the oxidation of free sugars to form glyoxal, methylglyoxal, and 3-deoxyglucosone. The final stage of this glycation is characterized by the formation of advanced glycation end products (AGEs).
It has been proposed that glycation-related modifications of human serum albumin (HSA), the main protein in serum, can alter the binding of this protein with drugs and small solutes [14-18]. HSA is the major carrier protein for many drugs and hormones in the circulation [19], which makes studies of HSA glycation of great potential interest in understanding how the role of this protein as a binding and transporting agent may be altered during diabetes. A few previous studies have been conducted to determine the levels of modification that occur at various regions on glycated HSA, but most of these reports have only looked at specific modifications such fructosyl-lysine (FL) or some types of AGEs (e.g., those formed from methylglyoxal) [19-22]. However, it is known there are actually many glycation-related modifications that can occur on HSA [19,20,22]. It has also been found that different levels of glycation can alter the binding properties of this protein [14,22-24]. These results indicate that there is a need for a better understanding of how the degree and types of glycation-related modifications on HSA may change as the extent of glycation for this protein is varied [1,20].
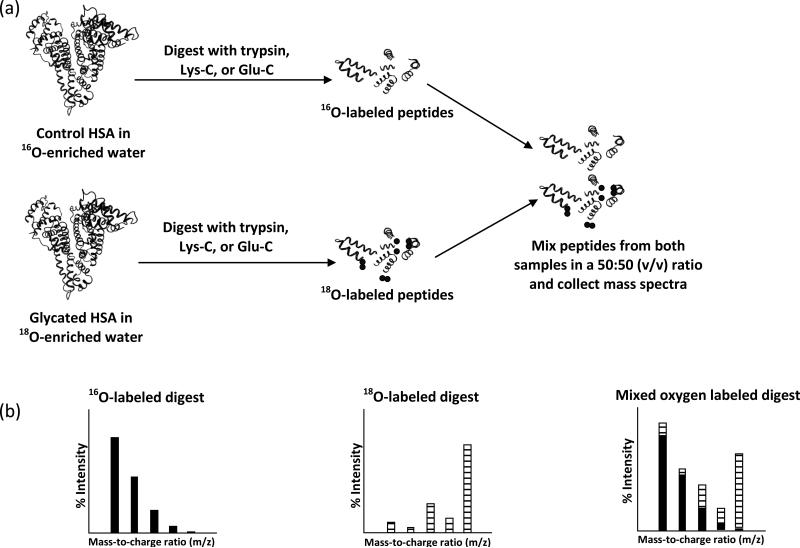
The purpose of this study is to quantitatively examine the modification patterns that occur for in vitro glycated HSA at various overall levels of glycation. These patterns will be examined by using 16O/18O-labeling and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). To analyze the glycation patterns, a control digest of HSA will be made in the presence of an 16O-enriched buffer, while similar digests of glycated HSA samples will be prepared in an 18O-enriched buffer (Fig. 2) [25-27]. These samples will then be mixed and the 16O/18O ratios for the peptides in each digest sample versus the control will be determined by using MALDI-TOF MS. The use of 18O-labeling in mass spectrometry has been shown to be highly sensitive to changes in peptide concentration, with detection limits in the femtomol range having been reported in Ref. [28]. MALDI-TOF and 16O/18O-labeling has previously been used to rank modification sites in a single, commercial sample of mildly glycated HSA [27] but has not yet been used to compare multiple samples of glycated HSA.
Figure 2.
(a) Method used for labeling peptides with 18O. HSA is reconstituted in buffer that is enriched with 16O-labeled water and glycated HSA is reconstituted in buffer that was enriched with 18O-labeled water. The HSA and glycated HSA samples are then digested in an identical fashion. The resulting peptide solution is then mixed in a 50:50 (v/v) fashion. (b) Mass spectra are collected for the 16O, 18O, and mixed oxygen labeled digests (left to right), and the relative abundances obtained from these spectra are used to determine the 16O/18O ratios. The solid bars represent the contribution of 16O and the hashed bars show the contribution of 18O to each m/z value in an isotope cluster.
In this current study, 16O/18O-labeling and MALDI-TOF MS will be used to determine which regions on HSA have undergone a change in their level of modification during glycation and to examine these changes as a function of reaction time or glycation conditions. A recent report has used qualitative peptide mass fingerprinting to examine the same glycated HSA samples that will be examined in this current study [29]; this earlier report found that several residues on glycated HSA can contain early or advanced glycation products (e.g., lysine 93, 199, 276, 281, 286,414, 439 or 524/525, and arginine 98, 197 or 521). It has also been observed in this prior work that some of these modifications occur at or near the major drug binding regions on HSA [29]. This current study will expand on these findings by providing quantitative information on the relative degree of modification that occurs at such regions of HSA and on how the pattern of these modifications can change over the time allowed for glycation. This work will be carried out by using in vitro samples of glycated HSA that are prepared under conditions mimicking those found in blood during diabetes. The information generated in these experiments should be valuable in understanding how changes in glycation may affect the ability of HSA to bind drugs and small solutes during diabetes. The methods described in this report should also be useful for the quantitative analysis and comparison of other proteins or protein modifications that are of interest to clinical and biomedical research.
2. Materials and methods
2.1. Materials
The following chemicals were purchased for Sigma-Aldrich (St. Louis, MO): des-Argbradykinin (97% pure), glu-fibrinopeptide (97%), angiotensin I (97%, acetate salt), 2,3-dihyroxybenzoic acid (98%), α-cyano-4-hydroxycinnamic acid (99%), HSA (99%, essentially fatty acid and globulin free), trypsin (sequence grade), Glu-C (sequence grade), Lys-C (sequence grade), D-glucose (99%), iodoacetamide (99%), trifluoroacetic acid (98%), sodium azide (99%), formic acid (96%), molecular biology-grade water (DNase, RNase, and protease free), methanol (HPLC-grade, 99.9%), 18O-enriched water (97%), 16O-enriched water (99.99%), and acetonitrile (HPLC-grade, 99.9%). Unless indicated otherwise, all water used for sample pretreatment and the preparation of solutions was obtained from a Nanopure system (Barnstead, Dubuque, IA).
2.2. Apparatus
The siliconized low retention microcentrifuge tubes (0.6 ml and 1.5 ml) and 0.20 μm nylon filters were obtained from Fisher Scientific (Pittsburgh, PA). Slide-A-Lyzer dialysis cassettes (7000 Da MW cutoff, 0.1-0.5 ml and 0.5-3 ml) were from Pierce (Rockford, IL). The μ-C18 ZipTip pipette tips (0.2 μl bed volume) were purchased from Millipore (Billerica, MA). Mass spectra were collected on a Voyager 6148 MALDI-TOF MS system (Applied/Perspective Biosystems, CA). The instrument settings were as follows: positive-ion delayed extraction reflection mode; delay time, 100 ns; accelerating voltage, 25 kV; guide wire voltage, 0.008% of accelerating voltage; grid voltage, 77% of accelerating voltage. Matlab 2009a, which included the bioinformatics toolbox, was obtained from Mathworks (Natick, MA). Mascot Wizard was obtained from Matrix Science (London, UK) [30].
2.3. Preparation and digestion of glycated HSA
The preparation of the glycated HSA has been described in detail in a previous report [29]. The HSA was dissolved at a concentration of 42 g/l (or 0.63 mmol/l) in pH 7.4, 0.2 mol/l potassium phosphate buffer. This buffer also contained 1 mmol/l sodium azide as a preservative and prepared using molecular biology-grade water [31,32]. The initial concentration of HSA was selected to mimic the level of HSA that is typically found in serum (i.e., 35–50 g/l). Two initial solutions of HSA were prepared: the first solution contained 15 mmol/l D-glucose (or 270 mg/dl), which represented a glucose concentration that is typically found in blood during poorly-controlled diabetes [33]. The second HSA solution was used as a control and did not contain glucose. The control solution was incubated for 5 weeks at 37 °C. The HSA/glucose mixture was incubated under the same conditions, with aliquots being removed after 2 weeks and 5 weeks. Following the incubation step, the HSA was removed from the other solution components by using size-exclusion chromatography and several cycles of dialysis against water. The purified HSA was lyophilized and stored at 4 °C until later use. The glycation content of each HSA sample was determined by using a modified fructosamine assay [29].
The pretreatment of HSA (i.e., denaturation, reduction, and alkylation) was performed according to recent methods reported in the literature [27,29]. Following these pretreatment steps, each HSA sample was placed into a 0.1–0.5 ml dialysis cassette. Dialysis was performed on these samples versus three 1000 ml portions of water for 4 h each at room temperature. The resulting control and glycated HSA solutions were adjusted to the same volume. A 40 μl aliquot of each HSA solution was placed into several low retention microcentrifuge tubes and these samples were dried under vacuum with no heating. The vials, which now contained approximately 80 μg of either the control HSA or glycated HSA, were stored at -80 °C until use. Digests of these samples were prepared using trypsin, Glu-C and Lys-C, as described previously [27]. The digested samples were stored at -80 °C until use.
2.4. MALDI-TOF MS
The procedures used for preparing the MALDI matrix, calibrating the mass spectrometer, and carrying out peptide fractionation for the various digests of HSA have been described in detail elsewhere [27,29]. The MALDI matrix consisted of a mixture of 2,3-dihyroxybenzoic acid and α-cyano-4-hydroxycinnamic acid. Glu-fibrinopeptide (1570.68 Da), des-Arg-bradykinin (904.47 Da), and angiotensin I (1296.68 Da) were used as external mass calibrants. The HSA digests were fractionated using μ-C18 ZipTip pipette tips and step gradients of 5, 10, 20, 30, or 50% acetonitrile in water [34-36]. The eluted peptides were mixed with the MALDI matrix, spotted onto a MALDI plate, dried, and analyzed by MALDI-TOF MS. Peptide mass mapping was performed with a maximum allowed mass error of 50 ppm.
The 16O/18O ratio for a given peptide in a mixed digest was obtained from the MALDI-TOF MS data by using Eqn. (1) [27,35].
| (1) |
The terms I’ and I” in Eqn. (1) represent the contributions from the 16O- and 18O-labeled components of the mixed digest, respectively. The subscripts 0, 2, and 4 in Eqn. (1) refer to whether the given contribution comes from peptides that contained zero (M+0 peak), one (M+2 peak), or 2 (M+4 peak) 18O-labels. These contributions were determined algebraically by incorporating mass/area values from both the 16O- and 18O-labeled spectra, as described earlier [27,35]. Unless stated otherwise, the measurement of the 16O/18O ratio for a given peptide in a collected fraction from a digest was performed in triplicate, and the 16O/18O ratios for the same peptide found in different fractions were combined and averaged. A Student's t-test [37] was carried out at the 90% confidence level to identify peptides with 16O/18O ratios in a test sample that were significantly higher than results for the same peptides in the reference, control sample.
3. Results and discussion
3.1. Initial analysis of glycated HSA
The control HSA sample used in this study had only a small amount of glycation-related modifications (i.e., less than 0.1 mol hexose per mol of HSA), as determined by a fructosamine assay [29]. The glycated HSA samples that were prepared using 2 or 5 weeks of incubation with 15 mmol/l glucose contained 0.7 (± 0.1) or 1.2 (± 0.1) mol hexose per mol of HSA when measured by the same approach. These levels of glycation represent the amount that is typically found in pre-diabetes (~0.5-1 mol hexose per mol HSA) or diabetes (1-15 mol hexose per mol HSA), respectively [38,39]. When combining the results for all digests, these samples were found to have usable sequence coverages of 86%, 82%, and 86%, respectively, for the quantitative studies that are described in this report (see Figure 3). The coverage obtained in this work was sufficient for the analysis of the N-terminus, all arginines, and 84-85% of the lysine residues on HSA in all of the samples that were examined. Analysis of the remaining 15-16% lysines in HSA was not required in this report because these particular residues have been found in prior studies to not have any measurable levels of glycation-related modifications [19-22].
Figure 3.
The usable sequence coverages for the samples examined in this study. The sequence coverage for the control HSA and five week glycated HSA sample is shown in bold and caps (Note: The coverages for these two samples were identical). The sequence coverage for the HSA-2 sample is shown by an underline. The lower case letters represents the remainder of the sequence in HSA, for which no 16O/18O ratio determinations were made.
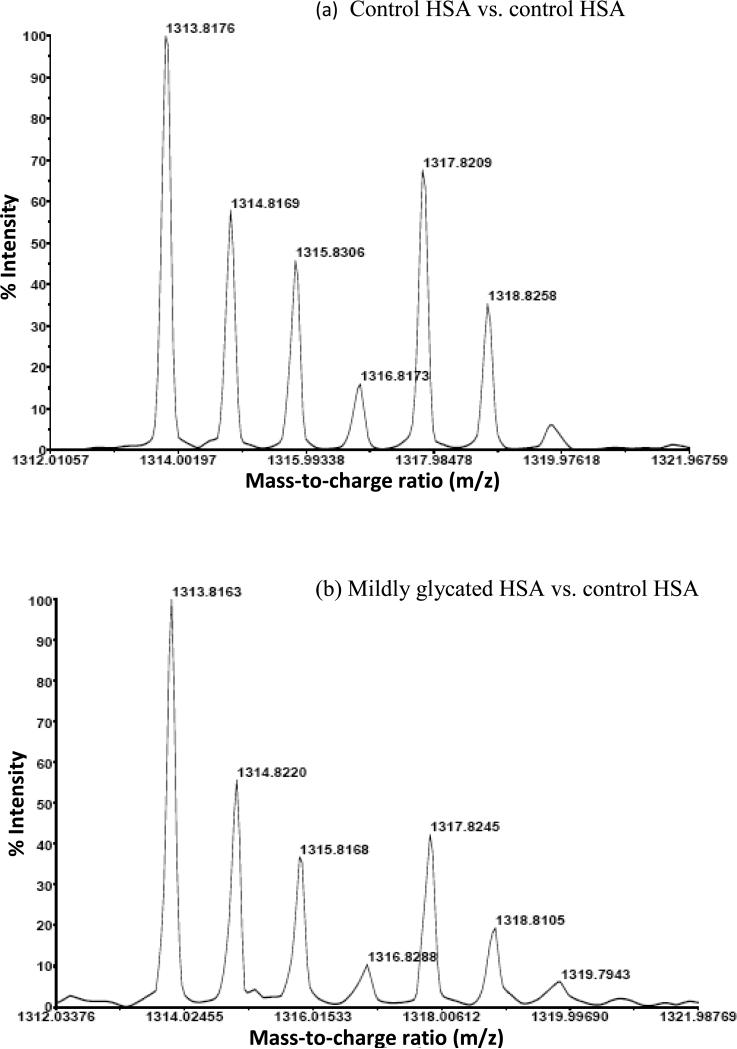
In the analysis method that was used in this study, the glycation or modification of a given peptide should have led to a decrease in the amount of unmodified peptide and an increase in the corresponding 16O/18O ratio for this region when comparing the control sample to a glycated HSA sample. Figure 4 illustrates this approach by showing some typical mass spectra that were obtained when there was an increase in the 16O/18O ratio for a peptide containing residues 521-531 of HSA. This particular region of HSA contains K525, which is a well-characterized glycation site on this protein [20]. Based on an analysis of the mass spectrum in Figure 4(a) and through the use of Eqn. (1), the reference 16O/18O ratio for this peptide in the original control sample was found to be 1.15 (± 0.13). When using the data in Figure 4(b), the 16O/18O ratio increased to 1.80 (± 0.46) in the mildly glycated HSA sample that was obtained after 2 weeks of incubation with glucose. An even larger increase in this ratio to 2.00 (± 0.46) was found in the glycated HSA sample after 5 weeks of incubation with glucose (data not shown). By using these ratios, it was possible to obtain a quantitative measure of the relative degree to which this and other regions of HSA were affected by glycation-related modifications at different incubation times with glucose.
Figure 4.
Mass spectra showing the change in 16O/18O ratio for a peptide from residues 521-531 of HSA as the level of glycation for HSA was increased in going from (a) the control digest to (b) glycated HSA prepared after two weeks of incubation with glucose. An even larger change was seen in this case when using the glycated HSA that was prepared after five weeks of incubation with glucose (data not shown). The region of HSA that was examined in this case contains K525, which is known to be a major glycation site on HSA [20].
To make it easier to compare changes in the 16O/18O ratios for different peptides from the glycated HSA samples versus normal HSA, a correction was also made for the small variations that occurred in the degree of 16O- or 18O-labeling during the production of peptide fragments from HSA. For instance, the 16O/18O ratios for peptides in the control sample (i.e., as found by examining the control sample versus itself when prepared in 16O-enriched or 18O-enriched buffer) varied from 0.58–2.76, 0.50–3.37, and 0.17–2.12 for the trypsin, Glu-C, and Lys-C digests, respectively. The variations seen in these values for the control most likely reflect the differences in how the given enzymes interact with various regions of HSA, as has been noted when using a similar approach to compare immobilized HSA versus soluble HSA [35]. In this type of situation, a comparison of the 16O/18O ratio for a test sample versus the same ratio for an appropriate control sample was found to be a more effective route for comparing the amount of modification that was occurring at different regions of HSA. This approach was employed in this study by determining 16O/18O ratios for 1) each glycated HSA sample versus a control sample of normal HSA and 2) the control HSA sample versus itself after digestion of this sample in both 16O-enriched and 18O-enriched water. A value referred to here as the “glycated/control index” was then determined for each peptide by dividing the 16O/18O ratio for that peptide in the glycated HSA/control HSA mixture by the 16O/18O ratio for the same peptide in the control HSA/control HSA mixture. The glycated/control index provided a normalized scale in which all peptides in the original control sample had an initial reference index of one. An index value greater than one was produced when a glycation-related modification occurred in a given region of HSA, with the size of this index increasing as the amount of modification increased. This feature made it possible with this index to better determine which regions of HSA were most affected by glycation-related modifications and to compare the time course of these processes, as will be discussed later.
Table 1 shows the 16O/18O ratios that were obtained for all of the peptides that were examined in this work and the glycated/control index values that were calculated for the various regions on HSA from which these peptides originated. The standard error of the mean for each 16O/18O ratio and for each calculated glycated/control index is also provided in Table 1. Regions on HSA that had significant levels of modification were identified by using a Student's t-test to compare the 16O/18O ratios for the control sample to the 16O/18O ratios for the glycated HSA samples. Peptides from the glycated HSA samples that gave an increase in the 16O/18O ratio versus the control sample at the 90% confidence level were considered to represent significant levels of modification. The corresponding glycated/control index values for these peptides were then used in further work to compare the levels of modification that were measured for peptides from different regions of HSA.
Table 1.
Results based on 16O/18O measurements for glycated HSA samples after two or five weeks of incubation in 15 mM glucosea
| 16O/18O Ratio, Number of Replicates (n) | Glycated/Control Index | |||||||
|---|---|---|---|---|---|---|---|---|
| Mass (u) | Error (ppm) | Residues | Control Sample | 2 Week Sample | 5 Week Sample | 2 Week Sample | 5 Week Sample | |
| Trypsin | 927.50 | 2 | 138-144 | 1.66 (± 0.08), 9 | 1.94 (± 0.14), 9 | 2.02 (± 0.14), 9 | 1.17 (± 0.13) | 1.22 (± 0.13) |
| 960.57 | 6 | 403-410 | 1.51 (± 0.10), 9 | 1.30 (± 0.06), 9 | 1.53 (± 0.08), 9 | 0.86 (± 0.09) | 1.01 (± 0.10) | |
| 1055.59 | -1 | 137-144 | 1.34 (± 0.13), 9 | 1.59 (± 0.18), 9 | 1.55 (± 0.07), 9 | 1.19 (± 0.19) | 1.16 (± 0.18) | |
| 1074.53 | -16 | 182-190 | 1.67 (± 0.31), 9 | 2.55 (± 0.25), 3 | 2.02 (± 0.65), 6 | 1.53 (± 0.58) | 1.21 (± 0.46) | |
| 1149.60 | -10 | 1-10 or 42-51 | 1.99 (± 0.33), 6 | 1.57 (± 0.33), 6 | 3.20 (± 0.69), 6 | 0.79 (± 0.27) | 1.61 (± 0.55) | |
| 1226.58 | -5 | 11-20 | 1.56 (± 0.24), 9 | 1.19 (± 0.16), 8 | 1.34 (± 0.13), 6 | 0.76 (± 0.17) | 0.86 (± 0.19) | |
| 1342.62 | -11 | 546-557 | 0.72 (± 0.07), 4 | 0.46 (± 0.05), 6 | 1.03 (± 0.09), 5 | 0.64 (± 0.11) | 1.43 (± 0.24) | |
| 1371.56 | -5 | 163-174 | 1.21 (± 0.12), 6 | 1.48 (± 0.13), 3 | 0.95 (± 0.13), 6 | 1.22 (± 0.24) | 0.79 (± 0.15) | |
| 1434.53 | -6 | 82-93 | 1.10 (± 0.10), 3 | 1.36 (± 0.31), 6 | 1.89 (± 0.38), 6 | 1.24 (± 0.39) | 1.72 (± 0.55) | |
| 1443.64 | -4 | 263-274 | 1.81 (± 0.19), 6 | 2.07 (± 0.54), 6 | 2.42 (± 0.37), 6 | 1.14 (± 0.36) | 1.34 (± 0.43) | |
| 1467.82 | -17 | 337-348 | 2.01 (± 0.18), 6 | 2.01 (± 0.41), 6 | 2.43 (± 0.26), 7 | 1.00 (± 0.25) | 1.21 (± 0.30) | |
| 1546.80 | 2 | 275-286 | 1.37 (± 0.23), 5 | 2.02 (± 0.24), 6 | 1.49 (± 0.33), 6 | 1.47 (± 0.44) | 1.09 (± 0.33) | |
| 1552.60 | -2 | 360-372 | 1.46 (± 0.19), 6 | 2.33 (± 0.27), 6 | 1.86 (± 0.08), 6 | 1.60 (± 0.29) | 1.27 (± 0.23) | |
| 1623.80 | 5 | 324-336 | 1.85 (± 0.03), 3 | 2.43 (± 0.15), 6 | 3.05 (± 0.16), 6 | 1.31 (± 0.11) | 1.65 (± 0.13) | |
| 1627.73 | 0 | 561-574 | 1.52 (± 0.14), 4 | 1.10 (± 0.20), 3 | 1.32 (± 0.23), 3 | 0.72 (± 0.19) | 0.87 (± 0.23) | |
| 1639.94 | 0 | 414-428 | 1.31 (± 0.08), 6 | 1.69 (± 0.19), 6 | 1.38 (± 0.08), 6 | 1.29 (± 0.18) | 1.05 (± 0.15) | |
| 1657.72 | -21 | 390-402 | 1.21 (± 0.24), 6 | 1.11 (± 0.12), 6 | 1.33 (± 0.24), 3 | 0.92 (± 0.26) | 1.10 (± 0.32) | |
| 1714.80 | 0 | 94-106 | 1.46 (± 0.07), 9 | 1.59 (± 0.05), 9 | 1.86 (± 0.13), 6 | 1.09 (± 0.10) | 1.27 (± 0.12) | |
| 1742.89 | -9 | 146-159 | 1.55 (± 0.09), 3 | 1.85 (± 0.20), 3 | 2.59 (± 0.43), 3 | 1.19 (± 0.25) | 1.67 (± 0.34) | |
| 1899.01 | 7 | 146-160 | 1.48 (± 0.19), 3 | 2.38 (± 0.56), 3 | 2.47 (± 0.24), 6 | 1.61 (± 0.46) | 1.67 (± 0.48) | |
| 1910.92 | -5 | 485-500 | 1.46 (± 0.10), 6 | 1.31 (± 0.07), 6 | 1.45 (± 0.06), 6 | 0.90 (± 0.09) | 0.99 (± 0.09) | |
| 2045.10 | 0 | 373-389 | 1.43 (± 0.05), 3 | 1.68 (± 0.00), 3 | 1.95 (± 0.17), 6 | 1.17 (± 0.11) | 1.36 (± 0.13) | |
| 2086.80 | -19 | 241-257 | 1.27 (± 0.07), 9 | 1.66 (± 0.10), 9 | 1.65 (± 0.08), 9 | 1.31 (± 0.12) | 1.30 (± 0.12) | |
| 2260.10 | 27 | 501-519 | 1.73 (± 0.27), 3 | 1.37 (± 0.33), 2 | 0.75 (± 0.14), 3 | 0.79 (± 0.27) | 0.43 (± 0.15) | |
| 2490.28 | -2 | 21-41 | 1.71 (± 0.10), 3 | 1.92 (± 0.20), 3 | 3.50 (± 0.42), 3 | 1.12 (± 0.19) | 2.05 (± 0.35) | |
| 2518.20 | -5 | 352-372 | 1.82 (± 0.17), 3 | 1.23 (± 0.31), 3 | 1.43 (± 0.39), 3 | 0.68 (± 0.26) | 0.79 (± 0.30) | |
| 2585.11 | -4 | 241-262 | 0.92 (± 0.03), 3 | 0.81 (± 0.02), 3 | 0.84 (± 0.06), 6 | 0.88 (± 0.08) | 0.91 (± 0.08) | |
| 2650.30 | 15 | 115-136 | 1.21 (± 0.02), 3 | 1.24 (± 0.07), 3 | 1.11 (± 0.01), 3 | 1.02 (± 0.06) | 0.92 (± 0.05) | |
| 2674.31 | -1 | 445-466 | 0.99 (± 0.03), 3 | 0.78 (± 0.05), 3 | 0.93 (± 0.21), 3 | 0.79 (± 0.19) | 0.94 (± 0.22) | |
| 2778.43 | 25 | 115-137 | 1.01 (± 0.01), 3 | 0.92 (± 0.02), 3 | 1.09 (± 0.05), 3 | 0.91 (± 0.05) | 1.08 (± 0.05) | |
| 2974.31 | -10 | 287-313 | 1.05 (± 0.04), 2 | Not detected | 1.21 (± 0.49), 2 | Not detected | 1.15 (± 0.47) | |
| Glu-C | 1031.45 | 3 | 369-376 | 1.92 (± 0.27), 6 | 1.44 (± 0.46), 3 | 1.43 (± 0.21), 6 | 0.75 (± 0.28) | 0.74 (± 0.28) |
| 1197.57 | -6 | 496-505 | 0.96 (± 0.34), 6 | 0.46 (± 0.05), 2 | 1.20 (± 0.27), 3 | 0.48 (± 0.21) | 1.25 (± 0.54) | |
| 1300.67 | 7 | 7-17 | 1.57 (± 0.18), 6 | 2.00 (± 0.41), 6 | 1.87 (± 0.24), 6 | 1.27 (± 0.34) | 1.19 (± 0.32) | |
| 1313.80 | 4 | 521-531 | 1.15 (± 0.13), 5 | 1.80 (± 0.46), 6 | 2.00 (± 0.46), 3 | 1.57 (± 0.57) | 1.74 (± 0.63) | |
| 1519.79 | 2 | 142-153 | 1.40 (± 0.09), 12 | 1.80 (± 0.23), 12 | 1.76 (± 0.06), 9 | 1.29 (± 0.19) | 1.26 (± 0.18) | |
| 1541.75 | 36 | 506-518 | 1.42 (± 0.23), 5 | 1.41 (± 0.39), 3 | 1.66 (± 0.09), 3 | 0.99 (± 0.33) | 1.17 (± 0.38) | |
| 1548.77 | -16 | 480-492 | 1.29 (± 0.07), 9 | 1.40 (± 0.05), 9 | 1.36 (± 0.11), 7 | 1.09 (± 0.11) | 1.05 (± 0.11) | |
| 1680.80 | -5 | 466-479 | 1.15 (± 0.10), 3 | 1.33 (± 0.22), 6 | 1.40 (± 0.29), 5 | 1.16 (± 0.33) | 1.22 (± 0.34) | |
| 1704.91 | -22 | 154-167 | 1.63 (± 0.08), 6 | 1.98 (± 0.03), 6 | 1.84 (± 0.13), 7 | 1.21 (± 0.11) | 1.13 (± 0.10) | |
| 1737.69 | -4 | 87-100 | 1.19 (± 0.14), 5 | 1.76 (± 0.29), 6 | 1.70 (± 0.20), 6 | 1.48 (± 0.34) | 1.43 (± 0.33) | |
| 1801.92 | 0 | 451-465 | 0.49, 1 | Not detected | 2.23, 1 | Not detected | 4.55 | |
| 1945.86 | -1 | 502-518 | 1.16 (± 0.02), 3 | 1.48 (± 0.04), 3 | 1.19 (± 0.20), 3 | 1.28 (± 0.21) | 1.03 (± 0.17) | |
| 1955.98 | -8 | 426-442 | 1.15 (± 0.04), 5 | 1.38 (± 0.05), 6 | 1.46 (± 0.04), 6 | 1.20 (± 0.07) | 1.27 (± 0.07) | |
| 2085.99 | -38 | 377-393 | 1.22 (± 0.10), 6 | 1.46 (± 0.11), 6 | 1.43 (± 0.14), 6 | 1.20 (± 0.18) | 1.17 (± 0.18) | |
| 2194.15 | -11 | 189-208 | 0.93 (± 0.05), 5 | 1.16 (± 0.40), 6 | 1.29 (± 0.22), 6 | 1.25 (± 0.49) | 1.39 (± 0.54) | |
| 2232.21 | -19 | 209-227 | 1.12 (± 0.04), 6 | 1.30 (± 0.09), 6 | 1.24 (± 0.03), 6 | 1.16 (± 0.10) | 1.11 (± 0.09) | |
| 2348.09 | -42 | 101-119 | 0.99 (± 0.02), 6 | 1.75 (± 0.45), 6 | 1.21 (± 0.05), 3 | 1.77 (± 0.46) | 1.22 (± 0.32) | |
| 2578.26 | -4 | 61-82 | 0.68 (± 0.01), 3 | 0.84 (± 0.03), 3 | 0.80 (± 0.06), 6 | 1.24 (± 0.10) | 1.18 (± 0.09) | |
| Lys-C | 854.46 | 6 | 206-212 | 0.51 (± 0.31), 3 | 1.40 (± 0.33), 3 | 1.76 (± 0.50), 5 | 2.75 (± 1.96) | 3.45 (± 2.47) |
| 973.52 | -1 | 5-12 | 1.13 (± 0.18), 3 | 0.93 (± 0.04), 3 | 1.04 (± 0.07), 5 | 0.82 (± 0.15) | 0.92 (± 0.17) | |
| 1149.62 | 4 | 1-10 | 1.00 (± 0.13), 3 | 1.64 (± 0.31), 6 | 1.99 (± 0.37), 3 | 1.64 (± 0.49) | 1.99 (± 0.59) | |
| 1342.63 | -5 | 546-557 | 1.48 (± 0.17), 3 | 1.10 (± 0.12), 3 | 1.07 (± 0.09), 3 | 0.74 (± 0.13) | 0.72 (± 0.13) | |
| 1352.76 | -3 | 403-413 | 1.29 (± 0.05), 9 | 1.09 (± 0.08), 9 | 1.32 (± 0.22), 12 | 0.84 (± 0.16) | 1.02 (± 0.19) | |
| 1371.55 | -10 | 163-174 | 1.44 (± 0.18), 6 | 1.43 (± 0.14), 6 | 1.23 (± 0.09), 6 | 0.99 (± 0.18) | 0.85 (± 0.15) | |
| 1443.63 | -8 | 263-274 | 1.37 (± 0.18), 7 | 1.36 (± 0.05), 6 | 1.28 (± 0.11), 6 | 0.99 (± 0.16) | 0.93 (± 0.15) | |
| 1529.86 | -7 | 213-225 | 1.27 (± 0.03), 6 | 1.26 (± 0.03), 6 | 1.21 (± 0.03), 6 | 0.99 (± 0.04) | 0.95 (± 0.04) | |
| 1552.59 | -4 | 360-372 | 1.31 (± 0.04), 6 | 1.71 (± 0.05), 6 | 1.40 (± 0.02), 6 | 1.31 (± 0.06) | 1.07 (± 0.05) | |
| 1657.74 | -9 | 390-402 | 1.54 (± 0.24), 5 | 1.48 (± 0.29), 6 | 1.34 (± 0.25), 5 | 0.96 (± 0.30) | 0.87 (± 0.27) | |
| 1714.79 | -7 | 94-106 | 1.17 (± 0.02), 9 | 1.25 (± 0.01), 9 | 1.17 (± 0.03), 6 | 1.07 (± 0.04) | 1.00 (± 0.03) | |
| 1924.05 | -17 | 415-432 | 0.73 (± 0.13), 3 | 0.52 (± 0.01), 3 | 0.39 (± 0.07), 3 | 0.71 (± 0.18) | 0.53 (± 0.13) | |
| 2045.05 | -22 | 373-389 | 1.28 (± 0.03), 3 | 1.17 (± 0.03), 3 | 1.30 (± 0.05), 3 | 0.91 (± 0.05) | 1.02 (± 0.05) | |
| 2052.15 | -17 | 414-432 | 1.26 (± 0.06), 6 | 1.01 (± 0.06), 6 | 1.15 (± 0.03), 6 | 0.80 (± 0.07) | 0.91 (± 0.08) | |
| 2259.99 | -16 | 21-41 | 1.53 (± 0.23), 6 | Not detected | Not detected | Not detected | Not detected | |
| 2348.99 | -20 | 74-93 | 0.83 (± 0.01), 3 | 0.80 (± 0.10), 6 | 0.95 (± 0.10), 6 | 0.96 (± 0.16) | 1.14 (± 0.19) | |
| 2490.24 | -17 | 21-41 | 0.79 (± 0.06), 3 | 0.65 (± 0.07), 3 | 0.98 (± 0.08), 3 | 0.82 (± 0.12) | 1.24 (± 0.19) | |
| 2585.09 | -11 | 241-262 | 0.93 (± 0.07), 9 | 0.88 (± 0.02), 6 | 0.92 (± 0.02), 3 | 0.95 (± 0.08) | 0.99 (± 0.08) | |
| 2674.29 | -11 | 445-466 | 0.88 (± 0.02), 3 | 0.86 (± 0.05), 3 | 0.70 (± 0.05), 3 | 0.98 (± 0.10) | 0.80 (± 0.08) | |
| 2807.45 | -6 | 138-159 | 0.84 (± 0.01), 3 | 0.84 (± 0.04), 3 | 0.75 (± 0.02), 3 | 1.00 (± 0.05) | 0.89 (± 0.05) | |
The values shown in parentheses represent ± 1 standard error of the mean. The values in bold represent cases for which the calculated 16O/18O ratio for the glycated sample was higher than the 16O/18O ratio for the control sample at the 90% confidence level.
3.2. Peptides with high 16O/18O ratios in mildly glycated HSA
A detailed examination was first made of the results for the mildly glycated HSA that was prepared using a 2 week incubation period with 15 mmol/l glucose. A total of 16 peptides in this sample gave 16O/18O ratios that were significantly higher (at the 90% confidence level) versus results for the same peptides in the control HSA sample. These peptides had a relative increase of 6% to 77% in their glycated/control indices versus the control. Based on these ratios, the largest amount of modification in the mildly glycated HSA sample was found to occur at residues 101-119, 1-10 or 42-51, 87-100, 360-372, 521-531, and 275-286. The peptides from these regions gave glycated/control indices that represented a relative increase of 47-77% in the 16O/18O ratios for these residues in the mildly glycated HSA versus the control sample. Smaller amounts of modification were found on residues 324-336, 414-428, 373-389, 241-257, 142-153, 154-167, 502-518, 426-442, 209-227, and 61-82 in the mildly glycated HSA, with glycated/control index values that represented an increase of 16-31% in the 16O/18O ratios versus the control sample.
The N-terminus, K525 and K281 are among the most common glycation sites that have been reported for HSA [19,20,27,34,40]. In this study, these residues were found to occur in peptides with glycated/control index values of 1.64, 1.57, and 1.47, respectively, for the mildly glycated HSA. These residues are all located in regions that were found to have the highest levels of modification in glycated HSA after 2 weeks of incubation with glucose. In addition to these regions, a peptide corresponding to residues 426-442 was found to have an elevated glycated/control index of 1.20 (± 0.07). The increase in the glycated/control index at this latter region was lower than that seen for the glycated/control index of the more commonly-identified modified regions in glycated HSA. However, this site is worth mentioning because it has been previously identified as a likely modification site for in vivo glycated HSA [20]. Residues 189-208 may also have had an elevated glycated/control index (e.g., due to modification at K199, a known glycation site on HSA) [20,34], with a value of 1.25 (± 0.49) for the mildly glycated HSA sample. While the variation in this index and the corresponding 16O/18O ratios meant that this increase was not significant at the 90% confidence level, this region did have a significant increase in its glycation/control index for the more highly glycated HSA that was produced after 5 weeks of incubation with glucose (see next section).
The regions in the mildly glycated HSA sample that were found to contain significant amounts of modification were compared to observations made in prior work with glycated HSA. For instance, all of the regions found to have moderate-to-high levels of modification in a previous report using a commercial in vitro preparation of minimally-glycated HSA [27] were also found to have moderate-to-high levels of modification for the mildly glycated HSA sample that was prepared and examined in this current study. These regions contained residues R81, K212/R218, R114, K525, and R428, which have all been noted to be important modification sites in glycated HSA [19,20,22,24,34].
Some other regions that were found to have measurable levels of modification in the mildly glycated HSA (e.g., residues 324-336, 241-257, 142-153, and 502-518) were unique to this current study and have not been previously reported. The possible modification sites in these regions include arginines 336, 257, 144 and 145. Although no modified peptides were found previously in these regions of HSA when using peptide mass fingerprinting [29], this may have been a result of the low levels of modifications seen in these regions in this current report (as indicated by 18O-labeling) and the variety of AGEs that could be formed as these sites. The binding of glucose or carbonyl-containing compounds at any of these sites has the potential to alter the structure and binding properties of HSA through allosteric effects. Furthermore, modified residues in regions 142-153 or 241-257 on HSA could potentially alter the binding properties of HSA because these residues are located in one of the major drug binding sites of this protein (i.e., Sudlow site I).
3.3. Peptides with high 16O/18O ratios in highly glycated HSA
A similar examination was made of the results for the more highly glycated HSA that was prepared after 5 weeks of incubation with 15 mmol/l glucose, as used here to represent typical levels of glycation that might be seen in diabetes. A total of 21 peptides in this sample were found to have a significant increase (at the 90% confidence level) in their glycated/control index values versus the control sample. These peptides gave an increase in the glycated/control index that ranged from 7% to 105% versus the control. The largest amounts of modification in this sample occurred within residues 21-41, 1-10 or 42-51, 521-531, 82-93, and 146-160 of HSA. These regions gave glycated/control ratios in the highly glycated HSA sample that were increased by 67 to 105% versus the control. Several additional residues containing lower amounts of modification were also identified, as listed in Table 1.
For the highly glycated HSA, peptides that contained the N-terminus or K525 gave the highest levels of modification. Other residues found to have significant levels of modification included K199, R114, R218, R428 and K439 (e.g., regions 189-208, 101-119, 209-227, and 426-442). All of these residues have previously been identified as being the most likely regions of modification on HSA when this protein is incubated with glucose [20,40] or methylglyoxal (i.e., a reactive dicarbonyl compound that leads to AGE formation) [22,24]. In addition, it was noted that the ranking of the major modification sites in the highly glycated HSA sample was similar to the ranking of modification sites for the mildly glycated HSA that was discussed in Section 3.2. For both of these samples, peptides containing the N-terminus had the highest amount of modification, followed by peptides that contained lysines 525, 199 and 439. Lysine 281, which was found to have a high glycated/control index in the mildly glycated HSA sample, had a glycated/control index in the highly glycated HSA sample that was also significantly elevated at the 90% confidence level.
3.4. Comparison of the glycated/control index for early and advanced glycation products
It was possible by combining the qualitative results from Ref. [29] with the quantitative data from this current study to determine the modifications that were associated with the greatest degrees of modification in the mildly and highly glycated HSA samples. For instance, in the mildly glycated HSA there were 5 peptides with elevated glycated/control index values, which corresponded to residues 1-10, 275-286, 87-100, and 414-428. The modifications that occurred in these general regions of HSA have been described in Ref. [29]. Such modifications included the formation of a dehydrate of fructosyl-lysine (FL-1H2O) at the N-terminus plus 1-alkyl-2-formyl-3,4-glycosyl-pyrrole (AFGP) at K4; the formation of FL at K276 plus FL-1H2O at K281; the formation of FL at K281 plus Nε-carboxyethyl-lysine (CEL) at K286 (or FL-2H2O at K281 plus pyrraline (Pyr) at K286); the production of FL-2H2O at K93 (or CEL at K93 plus Nε-(5-hydro-5-methyl-4-imidazolon-2-yl)ornithine (MG-H1) at R98); and the creation of FL-1H2O at K414 plus AFGP at R428 (or AFGP at K414 plus Nε-[5-(2,3,4-trihydroxybutyl)-5-hydro-4-imidazolon-2-yl]ornithine (3-DG-H1) at R428).
When these modified residues in the mildly glycated HSA were ranked in order of their glycated/control index values, the N+/K4 region had the highest amount of modification, followed by regions containing K276/K281/K286, R98/K93, and K414/R428. A peptide that corresponded to residues 196-205, and that was found earlier to contain FL at K199 plus 3-DGH1 at R197 [28], also had a relatively high glycated/control index, but the 16O/18O ratio for this peptide in the mildly glycated HSA sample was not significantly different from the control result at the 90% confidence level, as stated in Section 3.2. One general observation made from these results was that lysines (as opposed to arginines) had the highest amount of modification in the mildly glycated sample. It was also noted that FL, or closely-related dehydrated forms of FL, were found on these lysines. These observations are consistent with a model in which the formation of early glycation products is favored over AGEs at the relatively short incubation time of 2 weeks that was used to prepare the mildly glycated sample.
In the highly glycated HSA sample, peptides that were found to have elevated glycated/control index values included residues 521-531, 189-208, and 426-442. These regions of the highly glycated HSA have been determined in Ref. [29] to involve the formation of Pyr at K524 plus Nε-(5-hydro-4-imidazolon-2-yl)ornithine (G-H1) at R521; the creation of Pyr at K199 plus imidazolone B (IB) at R197; the formation of 3-DG-H1 or AFGP at R428; and the production of FL-1H2O at K525. When the modified residues were ranked in order of their glycated/control index values, the region containing K525, K524 and R521 was found to have the highest amount of modification, followed by the regions that contained K199/R197 and R428. A comparison of these results with those for the mildly glycated HSA indicated that the relative importance of AGEs versus early glycation products tended to increase with the time allowed for glycation. This finding is consistent with a model in which there is a gradual shift from early glycation products to these products plus AGEs over time and there is an increase in the complexity of the glycation patterns as a protein is allowed to react with glucose over longer time intervals [1].
3.5. Changes in the glycated/control index with overall level of glycation
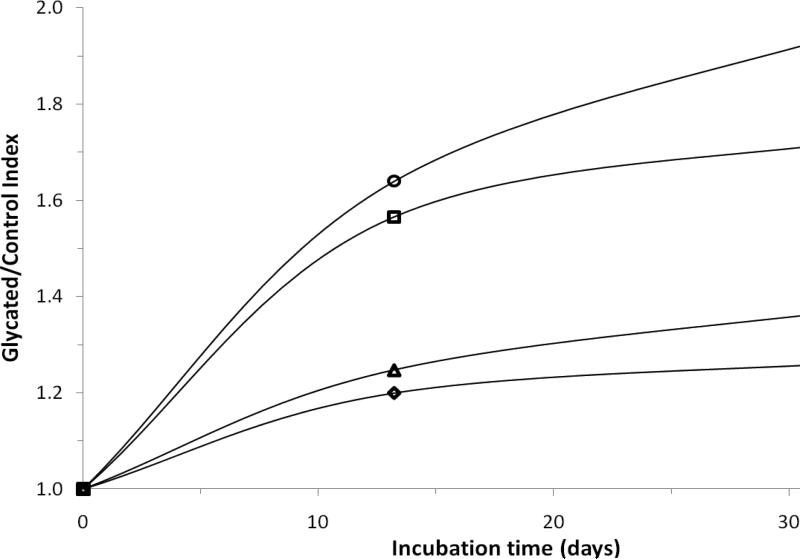
Most of the modified residues that were monitored in this study had a general increase in their glycated/control indices as the total level of glycation of HSA increased (e.g., as occurred when HSA was incubated over longer intervals of time with glucose). The majority of this increase occurred during the first 2 weeks of incubation between HSA and glucose. However, the rate of this increase did vary between different regions of HSA. This behavior is illustrated in Figure 5 for residues 1-10, 189-208, 521-531, and 426-442. As indicated earlier, these particular residues contain the N-terminus, K199, K525, and K439/R428, which are known to have high amounts of modification within both in vivo and in vitro glycated HSA [19,20,24]. There were also a few peptides that gave an increase in glycation-related modifications over 2 weeks of incubation with glucose, followed by an apparent reduction in the levels of modification at 5 weeks of incubation (i.e., see results for residues 101-119, 154-164, 275-286, 360-372, 414-428, and 502-518 in Table 1). However, the relative change in the modification levels between 2 and 5 weeks was generally small and/or not statistically significant in each these cases (Note: The only possible exception was the result for residues 360-372 in the Lys C digest, but no significant difference between the 2 and 5 week samples of glycated HSA was seen for the same peptide in the tryptic digest).
Figure 5.
Plot of the mean glycated/control index values versus time for the most commonly identified glycation sites on HSA [19,20,24]. These results are for residues 1-10 (○), 521-531 (□), 189-208 (Δ), and 426-442 (◇). Previous studies have indicated that the N-terminus, R521/K525, K199/K205 (the K205 being identified by tandem MS), and R428/K439 were modified to form mixtures of FL and AGEs [29]. The precision of the mean values shown in this plot are provided in Table 1.
Figure 5 shows that different regions of HSA did vary in their rates of modification due to glycation, which included related processes such as AGE formation. It was possible from this information to estimate the rate constants for these processes by preparing plots of the natural logarithm of the glycated/control index versus incubation time. This type of plot would be predicted to give a linear response if the glycation process followed a pseudo-first order model, with the slope being related to the rate constant for the modification process. A pseudo-first order model was sufficient in this case for use in the relative comparison of reaction rates and was reasonable because glucose was initially present in a 24-fold mole excess versus HSA (Note: additional time points would be needed to better determine the order and rates of these reactions in future studies). When the data in Figure 5 were examined according to a pseudo-first order model, the apparent rate constants for the initial rate of glycation product formation at the N-terminus, K525, K439/R428, and K199 (or K205) were estimated to be 2.5 × 10-5, 2.2 × 10-5, 1.1 × 10-5, and 1.1 × 10-5 min-1, respectively, based on data acquired over incubation times of up to 2 weeks at pH 7.4 and 37°C. The rate constants obtained using data for incubation times up to 5 weeks for the same sites were 1.3 × 10-5, 1.0 × 10-5, 0.7 × 10-5, and 0.6 × 10-5 min-1.
The other regions on HSA that were examined by this approach gave a similar range of rate constants for the glycation process. For instance, the modifications that occurred over 2 weeks at residues 241-257 (R257), 142-153 (R145), 61-82 (K64/K73), 154-167 (K159/R160), and 209-227 (R218) gave pseudo-first rate order constants for these reactions of 1.3 × 10-5, 1.2 × 10-5, 1.1 × 10-5, 0.9 × 10-5, and 0.7 × 10-5 min-1. Using data for the same sites over 5 weeks of incubation gave rate constants of 0.6 × 10-5, 0.6 × 10-5, 0.4 × 10-5, 0.3 × 10-5 and 0.3 × 10- 5 min-1. The results obtained over 2 weeks of incubation for residues 101-119 (R114), 360-372 (K372) and 275-286 (K281) gave rate constants of 2.8 × 10-5, 2.3 × 10-5, and 1.9 × 10-5 min-1, while data over 5 weeks of incubation resulted in rate constants of 0.8 × 10-5, 0.7 × 10-5 and 0.4 × 10-5 min-1. Although the rate of modification did vary from one region of HSA to the next, all of the observed regions and modifications gave pseudo-first order rate constants in the range of 10-6 to 10-5 min-1 under the incubation conditions used in this study.
3.6 Comparison of levels of modification for in vitro versus in vivo glycated HSA
In a recent study [41], FL modifications within in vivo glycated HSA were compared and measured in samples from 5 diabetic patients. These samples were processed by digesting the serum samples with trypsin, passing the digests through a phenylboronic acid column and analyzing the retained peptides by LC/MS/MS. The relative amount of each FL-modified peptide was then determined by comparing the signal to that for the FL-modified peptide with the largest response (i.e., a peptide containing K525). Table 2 compares the modification sites an relative ranking of these glycation sites with the results that were obtained for the same types of modifications in this current study.
Table 2.
Comparison of quantitative data obtained for in vitro glycated HSA and for in vivo glycated HSAa
| Change Relative to K525 | |||||
|---|---|---|---|---|---|
| Digest | Mass (u) | Residueb | 2 Week Sample | 5 Week Sample | In vivo HSA [41] |
| Trypsin | 1149.60 | 1-10 or K51c | 0.50 (1) | 0.93 (8) | 3.3 (2) |
| Lys-C | 2585.09 | K262 | 0.61 (2) | 0.57 (1) | 1.2 (1) |
| Glu-C | 1680.80 | K475 | 0.74 (3) | 0.70 (6) | 7.6 (4) |
| Trypsin | 2045.10 | K378 | 0.75 (4) | 0.78 (7) | 10.8 (5) |
| Glu-C | 1704.91 | K162 | 0.77 (5) | 0.65 (4) | 59.9 (10) |
| Glu-C | 2578.26 | K64 | 0.79 (6) | 0.68 (5) | 7.4 (3) |
| Trypsin | 1639.94 | K414 | 0.82 (7) | 0.60 (2) | 11.5 (6) |
| Trypsin | 1623.80 | K335 | 0.83 (8) | 0.95 (9) | 27.5 (8) |
| Trypsin | 1546.80 | K276 | 0.94 (9) | 0.63 (3) | 46.7 (9) |
| Glu-C | 1313.80 | K525 or K524/K525 | 1.00 (10) | 1.00 (10) | 100 (11) or 13.5 (7) |
| Lys-C | 1149.62 | 1-10 or K51c | 1.04 (11) | 1.14 (11) | 3.3 (2) |
The in vivo results are based on data from Ref. [41]. The values for the in vitro samples were determined by comparing the glycated/control index for the indicated peptide to the same index for a peptide containing K525. The relative ranking is given in parenthesis and is in increasing order from 1-13. The values in italics represent residues that have a similar modification rank in all three samples. Values in bold represent residues that have a similar rank in the two week in vitro glycated HSA and in vivo glycated HSA. The underlined values represent residues that are similarly ranked in the five week in vitro glycated HSA and in vivo glycated HSA.
The amino acid sequence for precursor HSA was used for peptide mapping in Ref. [41]. The listed sites were converted to those for the same amino acids in the sequence for mature HSA, as used in this current study, for the sake of consistency in the comparison.
This particular 16O/18O ratio could correspond to either residues 1-10 or 42-51. These peptides could not be differentiated at the 50 ppm level of mass accuracy that was used as the cut-off in this report.
Several peptides were found to have high levels of modification for both the in vitro glycated HSA in this current report and the in vivo glycated HSA in Ref. [41]. Of the 18 glycation sites that were found in Ref. [41], eleven of these sites were also associated with high and significant 16O/18O ratios in this current study. This group included all of the sites that were found to have the greatest levels of FL formation in Ref. [41]. Of the remaining FL sites that were examined previously [41], four were not included in the sequence coverage in this current study but contained only small levels of FL modification in the prior report (i.e., K181, K351, K541, and K545). One of the remaining FL sites (K233) has been found to be modified in prior work with in vitro glycated HSA [27]; this site may have been elevated in this current report, although the location of this modification could not be assigned definitively within a mass accuracy of 50 ppm. The remaining 2 sites that were monitored in Ref. [41] (i.e., K174 and K359) were also examined in this current study but were associated with low 16O/18O ratios. This latter result was in agreement with the relatively low levels of FL modification that were seen for the same regions in in vivo glycated HSA [41].
A comparison of the relative ranking of the detected FL sites in both the in vivo [41] and in vitro studies revealed that K525 contained the largest amount of modification, followed by lysines 335, 378, 475, and 262. A comparison of the in vivo glycated samples with the minimally glycated HSA that was prepared in vitro using 2 weeks of incubation with glucose revealed an even greater level of similarity that followed the relative pattern of modification K525 > K276 > K435 > K414 > K378 > K475 > K262. This result indicates that this in vitro glycated HSA sample was a reasonable model for the in vivo glycated HSA. A comparison of the same in vivo glycated HSA with the glycated HSA prepared in vitro after 5 weeks of incubation with glucose also compared well but with a slightly lower degree of similarity (K525 > K335 > K378 > K475 > K262).
The high level of similarity between the 2 week in vitro glycated HSA sample and the in vivo glycated HSA when looking at only FL adducts gives strong support to a model in which FL generation is more important than AGE formation at the early stages of glycation. A comparison of the 5 week in vitro glycated HSA and the in vivo glycated HSA further indicates that some alterations in the relative ranking of these modification sites can then occur as AGEs accumulate over time, coupled with the presence of some FL degradation. These results are all consistent with the results noted earlier in Section 3.4, in which the amounts of FL or AGE adducts were found to vary with the total extent of glycation and time allowed for glycation.
4. Conclusions
This paper investigated the use of 16O/18O-labeling and MALDI-TOF MS to provide quantitative information on the modification and modification patterns that occur on in vitro glycated HSA as a function of glycation time. The work used a glycated/control index to correct for variations in 16O/18O ratios within the control sample throughout the structure of HSA. This index also made it possible to compare the levels of modification from different regions of HSA, to examine the glycation patterns of HSA under various reaction conditions, and to estimate the rates of these modification reactions. The approach used in this report is not limited to HSA or glycation but could also be used to examine other types of modifications or protein systems.
By comparing the glycated/control index values, the largest levels of modification in HSA after 2 weeks of incubation with glucose occurred in regions 101-119, 1-10 or 42-51, 87-100, 360-372, 521-531, and 275-286. After 5 weeks of incubation, the largest amount of modification was found on residues 21-41, 1-10 or 42-51, 521-531, 82-93, and 146-160. Many of these regions contained residues that have been previously identified as major glycation sites on HSA, including the N-terminus, K199, K439, and K525 [20,40]. The glycation patterns found in this work supported a model in which FL and closely-related early glycation products dominated at short incubation times (e.g., 2 weeks), with the formation of AGEs also becoming important at longer incubation times (e.g., 5 weeks). Although the rates of these modifications varied from one region of HSA to the next, these processes were estimated to have pseudo-first order rate constants in the general range of 10-6 to 10-5 min-1.
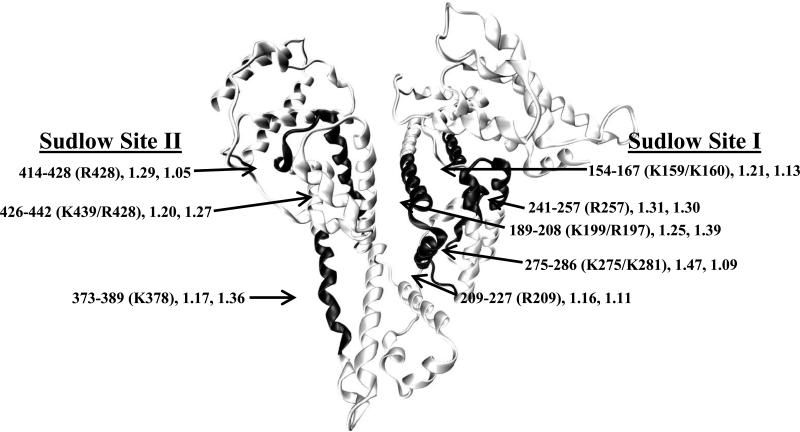
The physiological importance of these modifications can be illustrated by comparing the locations of the affected regions on HSA with the locations of Sudlow sites I and II (i.e., the major drug binding sites on this protein). As shown in Figure 6, and based on data or methods from Refs. [42,43], significant levels of modification were found to occur at or near both Sudlow sites I and II in the mildly glycated HSA and more highly glycated HSA samples. These modifications included those occurring at K199, K150 or K160, K275, K281, and R209 and R257, which are all located within Sudlow site I. Modifications at or near Sudlow site II included those involving K378 and K439 as well as R428. The location of these modifications explains why glycation has been observed to alter the binding of some drugs to Sudlow sites I and/or II of HSA [23,44,45]. The changes in the levels of these modifications and in the glycation pattern of HSA during the incubation of this protein with glucose helps explain why the binding of drugs at these sites has been found to change in the presence of different levels of glycation for HSA, as observed using samples comparable to those employed in this current report [44,45]. It was further found that the relative order of residues that formed early stage glycation products for the in vitro glycated HSA were similar to the order that has been recently reported for in vivo glycated HSA, especially when comparing the residues with the highest levels of modification [41]. Overall, this work should provide a more complete picture of how glycation affects the structure of HSA and the ability of the protein to transport and bind to drugs in individuals that are affected by diabetes. This information, in turn, should eventually lead to better methods of treatment for this disease as part of a regime based on personalized medicine.
Figure 6.
Locations of regions on HSA that gave high values for the glycated/control index and that occurred near Sudlow sites I and II. This image was generated using the crystal structure of HSA [41] and Visual Molecular Dynamics software [42]. This structure includes information on the region of HSA that is being modified, the specific residues that were most likely modified in this region [34], and the glycated/control index values for this regions in the glycated HSA samples after two or five weeks of incubation.
Acknowledgments
This research was supported by the National Institute of Health (NIH) under grant R01 DK069629. Support for the remodeled facilities that were used to perform these experiments was provided by NIH grant RR015468-001. Mass spectrometry data were acquired in the Nebraska Center for Mass Spectrometry, which is supported by National Cancer Institute grant P30 CA36727, NIH grants P20 RR15635 and RR015468, and the Nebraska Research Initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thornalley PJ, Langborg A, Minhas HS. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem J. 1999;344:109–16. [PMC free article] [PubMed] [Google Scholar]
- 2.Horiuchi S. Advanced glycation end products (AGE)-modified proteins and their potential relevance to atherosclerosis. Trend Cardiovasc Med. 1996;6:163–168. doi: 10.1016/1050-1738(96)00050-3. [DOI] [PubMed] [Google Scholar]
- 3.Araki N, Ueno N, Chakrabarti B, Morino Y, Horiuchi S. Immunochemical evidence for the presence of advanced glycation end products in human lens proteins and its positive correlation with aging. J Biol Chem. 1992;267:10211–4. [PubMed] [Google Scholar]
- 4.Hodge JE. Chemistry of browning reactions in model systems. J Agric Food Chem. 1953;1:928–43. [Google Scholar]
- 5.Gavin JR, Alberti KGMM, Davidson MB, DeFronzo RA, Drash A, Gabbe SG, et al. Report on the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 2002;25:S5–20. doi: 10.2337/diacare.26.2007.s5. [DOI] [PubMed] [Google Scholar]
- 6.Lapolla A, Fedele D, Reitano R, Bonfante L, Guizzo M, Seraglia R, et al. Mass spectrometric study of in vivo production of advanced glycation end-products/peptides. J Mass Spectrom. 2005;40:969–72. doi: 10.1002/jms.842. [DOI] [PubMed] [Google Scholar]
- 7.Lapolla A, Fedele D, Seraglia R, Traldi P. The role of mass spectrometry in the study of non-enzymatic protein glycation in diabetes: An update. Mass Spectrom Rev. 2006;25:775–97. doi: 10.1002/mas.20090. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Ames JM, Smith RD, Baynes JW, Metz TO. A perspective on the Maillard reaction and the analysis of protein glycation by mass spectrometry: Probing the pathogenesis of chronic disease. J Proteome Res. 2009;8:754–69. doi: 10.1021/pr800858h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nursten H. The Maillard Reaction. Royal Society of Chemistry; Cambridge, UK: 2005. [Google Scholar]
- 10.Monnier VM. Nonenzymatic glycosylation, the Maillard reaction and the aging process. J Gerontol. 1990;45:B105–11. doi: 10.1093/geronj/45.4.b105. [DOI] [PubMed] [Google Scholar]
- 11.Stitt AW, Curtis TM. Advanced glycation and retinal pathology during diabetes. Pharmacol Rep. 2005;57:156–68. [PubMed] [Google Scholar]
- 12.Ahmed N, Argirov OK, Minhas HS, Cordeiro CAA, Thornalley PJ. Assay of advanced glycation endproducts (AGEs): Surveying AGEs by chromatographic assay with derivitization by 6-aminoquinolyl-N-hydroxysuccinimidyl-carbamate and application to Nε-carboxymethyl-lysine- and Nε-(1-corboxyethyl)lysine-modified albumin. Biochem J. 2002;364:1–14. doi: 10.1042/bj3640001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thornalley PJ. Dicarbonyl intermediates in the Maillard reaction. Ann NY Acad Sci. 2005;1043:111–7. doi: 10.1196/annals.1333.014. [DOI] [PubMed] [Google Scholar]
- 14.Barzegar A, Moosavi-Movahedi AA, Sattarahmady N. Spectroscopic studies of the effects of glycation of human serum albumin on L-trp binding. Protein Pept Lett. 2007;14:13–8. doi: 10.2174/092986607779117191. [DOI] [PubMed] [Google Scholar]
- 15.Vorum H, Fisker K, Otagiri M, Pedersen AO, Hansen UK. Calcium ion binding to clinically relevant chemical modifications of human serum albumin. Clin Chem. 1995;41:1654–61. [PubMed] [Google Scholar]
- 16.Koyama H, Sugioka N, Uno A, Mori S, Nakajima K. Effects of glycosylation of hypoglycemic drug binding to serum albumin. Biopharm Drug Dispos. 1997;18:791–801. doi: 10.1002/(sici)1099-081x(199712)18:9<791::aid-bdd66>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Shaklai N, Garlick RL, Bunn HF. Nonenzymatic glycosylation of human serum albumin alters its confirmation and function. J Biol Chem. 1984;259:3812–7. [PubMed] [Google Scholar]
- 18.Mandula H, Parepally JMR, Feng R, Smith QR. Role of site-specific binding to plasma albumin in drug availability to brain. J Pharmacol Exp Therapeut. 2006;317:667–75. doi: 10.1124/jpet.105.097402. [DOI] [PubMed] [Google Scholar]
- 19.Peters T. All about Albumin: Biochemistry, Genetics, and Medical Applications. Academic Press; San Diego: 1996. [Google Scholar]
- 20.Iberg N, Fluckiger R. Nonenzymatic glycosylation of albumin in vivo. J Biol Chem. 1986;261:13542–5. [PubMed] [Google Scholar]
- 21.Capote FP, Scherl A, Müller M, Waridel P, Lisacek F, Sanchez JC. Glycation isotopic labeling (GIL) with 13C-reducing sugars for quantitative analysis of glycated proteins in human plasma. Mol Cell Proteomics. 2010;9:579–97. doi: 10.1074/mcp.M900439-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed N, Dobler D, Dean M, Thornalley PJ. Peptide mapping identifies hotspot site of modification in human serum albumin by methylglyoxal involved in ligand binding and esterase activity. J Biol Chem. 2005;280:5724–32. doi: 10.1074/jbc.M410973200. [DOI] [PubMed] [Google Scholar]
- 23.Joseph KS, Hage DS. The effects of glycation on the binding of human serum albumin to warfarin and L-tryptophan. J Pharm Biomed Anal. 2010;53:811–8. doi: 10.1016/j.jpba.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed N, Thornalley PJ. Peptide mapping of human serum albumin modified by methylglyoxal in vitro and in vivo. Ann NY Acad Sci. 2005;1043:260–6. doi: 10.1196/annals.1333.031. [DOI] [PubMed] [Google Scholar]
- 25.Stewart II, Thomson T, Figeys D. 18O labeling: A tool for proteomics. Rapid Comm Mass Spec. 2001;15:2456–65. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 26.Schnolzer M, Jedrzejewski P, Lehmann WD. Protease-catalyzed incorporation of 18O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis. 2005;17:945–53. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 27.Barnaby OS, Wa C, Cerny RL, Clarke W, Hage DS. Quantitative analysis of glycation sites on human serum labeling using 16O/18O labeling and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chim Acta. 2010;411:1102–10. doi: 10.1016/j.cca.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bantscheff M, Dümpelfeld B, Kuster B. Femtomol sensitivity post-digest 18O labeling for relative quantification of differential protein complex composition. Rapid Commun Mass Spectrom. 2004;18:869–76. doi: 10.1002/rcm.1418. [DOI] [PubMed] [Google Scholar]
- 29.Barnaby OS, Cerny RL, Clarke W, Hage DS. Comparison of modification sites formed on human serum albumin at various stages of glycation. Clin Chim Acta. 2011;412:277–285. doi: 10.1016/j.cca.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Atanasova M, Konova E, Betova T, Baydanoff S. Non-enzymatic glycation of human fibrillin-1. Gerontology. 2009;55:73–81. doi: 10.1159/000157436. [DOI] [PubMed] [Google Scholar]
- 32.Mitsuhashi T, Li YM, Fishbane S, Vlassara H. Depletion of reactive advanced glycation endproducts from diabetic uremic sera using a lysozyme-linked matrix. J Clin Invest. 1997;100:847–54. doi: 10.1172/JCI119600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner RC, Cull CA, Frighi V, Holman RR. Glycemic control with diet, sulfonylurea, metformin, or insulin in patients with type 2 diabetes mellitus. J Am Med Assoc. 1999;281:2005–12. doi: 10.1001/jama.281.21.2005. [DOI] [PubMed] [Google Scholar]
- 34.Wa C, Cerny RL, Clarke WA, Hage DS. Characterization of glycation adducts on human serum albumin by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chim Acta. 2007;385:48–60. doi: 10.1016/j.cca.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wa C, Cerny RL, Hage DS. Identification and quantitative studies of protein immobilization sites by stable isotope labeling and mass spectrometry. Anal Chem. 2006;78:7967–77. doi: 10.1021/ac0609935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wa C, Cerny R, Hage DS. Obtaining high sequence coverage in matrix-assisted laser desorption time-of-flight mass spectrometry for studies of protein modification: analysis of human serum albumin as a model. Anal Biochem. 2006;349:229–41. doi: 10.1016/j.ab.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Meier PC, Zund RE. Statistical Methods in Analytical Chemistry. Wiley; New York: 1993. [Google Scholar]
- 38.Abordo EA, Thornalley PJ. Pro-inflammatory cytokine synthesis by human monocytes induced by proteins minimally-modified by methylglyoxal. In: O'Brien J, Nursten HE, Crabbe MJC, Ames JM, editors. The Maillard Reaction in Foods and Medicine. The Royal Society of Chemistry; Cambridge, UK: 1998. pp. 357–62. [Google Scholar]
- 39.Lapolla A, Fedele D, Seraglia R, et al. A new effective method for the evaluation of glycated intact plasma proteins in diabetic subjects. Diabetologia. 1995;38:1076–81. doi: 10.1007/BF00402178. [DOI] [PubMed] [Google Scholar]
- 40.Robb DA, Olufemi SO, Williams DA, Midgley JM. Identification of glycation at the N-terminus of albumin by gas chromatography-mass spectrometry. Biochem J. 1989;261:871–8. doi: 10.1042/bj2610871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frolov A, Hoffmann R. Identification and relative quantification of specific glycation sites in human serum albumin. Anal Bioanal Chem. 2010;397:2349–56. doi: 10.1007/s00216-010-3810-9. [DOI] [PubMed] [Google Scholar]
- 42.Sugio S, Kashima A, Mochizuki S, Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–46. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 43.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Joseph KS, Anguizola J, Hage DS. Chromatographic analysis of acetohexamide binding to glycated human serum albumin. J Chromatogr B. 2010;878:2775–81. doi: 10.1016/j.jchromb.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph KS, Anguizola J, Hage DS. Binding of tolbutamide to glycated human serum albumin. J Pharm Biomed Anal. 2011;54:426–32. doi: 10.1016/j.jpba.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]