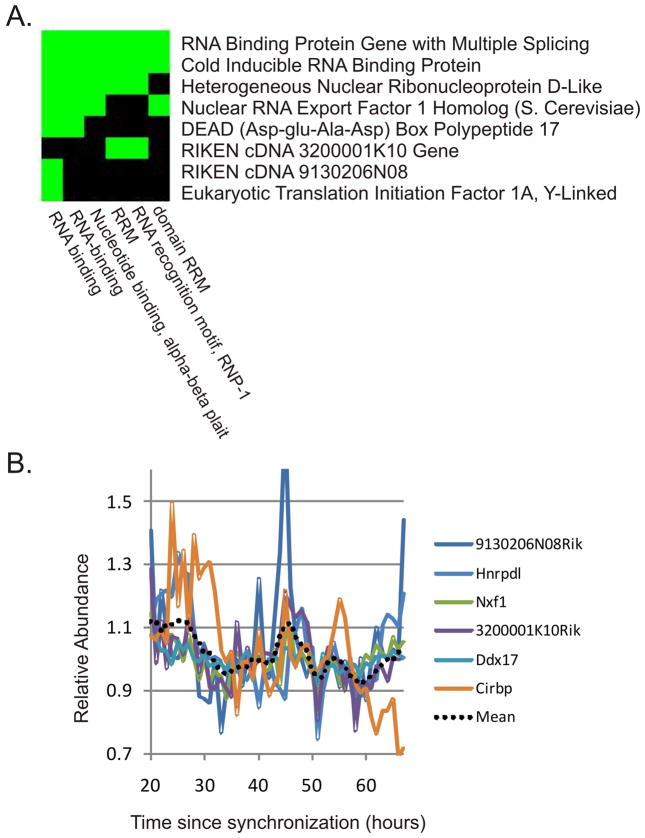
Figure 5. DAVID analysis of cycling genes detected by JTK_CYCLE in NIH3T3 cells reveals a cluster of RNA-binding genes with similar phases.
JTK_CYCLE was used to re-analyze cycling transcripts in synchronized NIH3T3 cells (Hughes et al. 2009). JTK_CYCLE detected more than twice as many cycling transcripts (N=30) as previously reported. DAVID analysis was performed on these 30 transcripts to detect enriched functional classes. Among the most enriched groups was a cluster of eight genes involved in RNA binding and recognition (A). Green blocks indicate that the annotations on the x-axis are present in the genes on the y-axis. Of these eight genes, six show similar phases of their oscillations (B), suggesting a common underlying mechanism (the dotted line represents a moving average of the median-normalized expression patterns of all six transcripts).