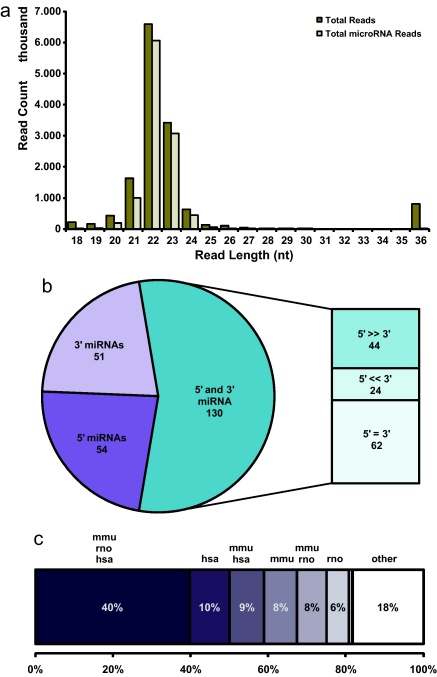
Fig. 2.
Hairpin classification and Chinese hamster ovary miRNA conservation. (a) Bar chart showing total read counts over read length for the complete read set (dark) compared to reads that had mapped the comprehensive miRNA genome and can therefore be considered as conserved miRNA reads (bright). (b) Of 235 canoncial miRNA hairpins that were discovered in CHO cells, 105 miRNA had been mapped at either the 5′ (54) or 3′ (51) position, while 130 hairpins had been mapped at both hairpin arms. The ratio of 5′ and 3′ read abundances was calculated for these 130 hairpins, resulting in 44 instances where the 5p/3p ratio exceeded an arbitrary ratio cut-off of 20:1, while in 24 instances it was below 1:20. (c) Out of 224 miRNAs that showed perfect identity to miRBase miRNA sequences, 82% had a human, mouse, or rat ortholog. Among the remaining 18% that did not have a perfect human, or a rodent ortholog, cow, platypus, and chicken were the most frequently found species.