Figure 8.
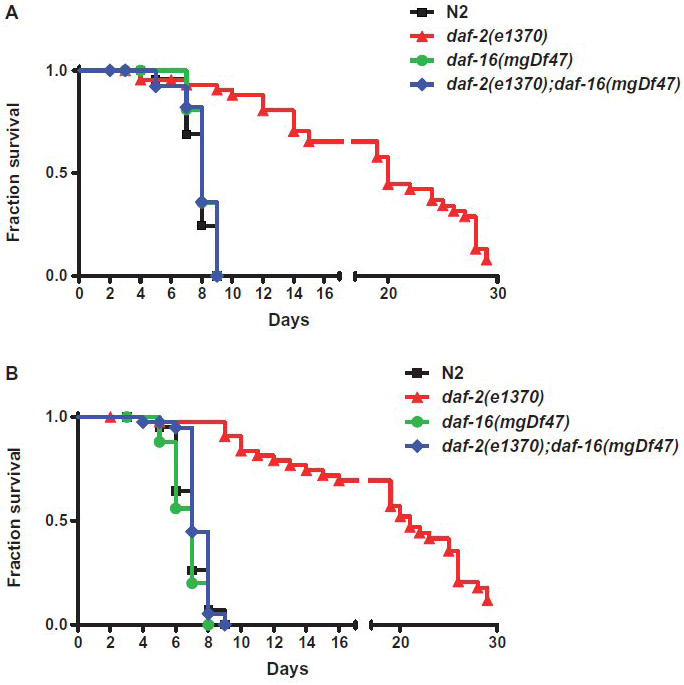
Survival of pathogen and stress-resistant C. elegans fed Legionella. (A) Kaplan-Meier survival plot of C. elegans N2 (squares, n=45), daf-2(e1370) (triangles, n=46), daf-16(mgDf47) (circles, n=46), and daf-2(e1370)/daf-16(mgDf47) (diamonds, n=45) fed L. pneumophila Philadelphia-1 type strain. (B) Kaplan-Meier survival plot of C. elegans N2 (squares, n=45), daf-2(e1370) (triangles, n=45), daf-16(mgDf47) (circles, n=27), and daf-2(e1370)/daf-16(mgDf47) (diamonds, n=41) fed on L. longbeachae type strain. P < 0.0001 by pairwise comparison by the log-rank test of daf-2 versus wild-type N2 for both Legionella strains. All other nematode strain pairwise comparisons versus wild-type N2 were found to be statistically insignificant. Data is representative of one of three independent experiments.
