Figure 7.
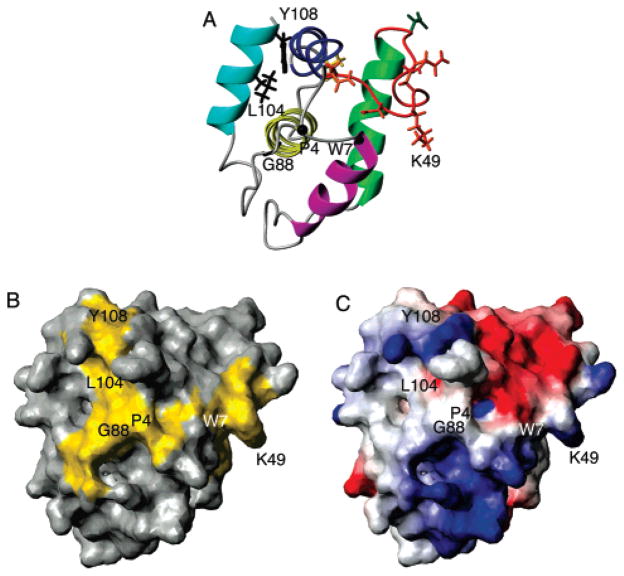
Residues affected by PIP2-C4 titration are mapped on the monomer structure of EIAV MA (A). Helices are colored magenta for Helix1, green for Helix2, blue for Helix3, yellow for Helix4, and cyan for Helix5. The loop between Helix2 and Helix3 is colored red. Strongly affected residues have their side chain bonds displayed. The backbone atoms are drawn for G88. Residues D42 and V63 are colored dark green and gold, respectively. Residues N44, E48, K49, D50, and Q52 are colored orange-red. Residues G88, L104, and Y108 are colored black. Residues P4 and W7 are also indicated with their Cα atoms drawn. The amide resonance for P4 is not available, while W7 shows a change in chemical shifts [just under the standard of deviation cutoff (Figure 5)], and they are in the PIP2 interaction site. (B) PIP2 interaction surface colored yellow using atoms from K49, G88, L104, Y108, P4, and W7. (C) Electrostatic surface plot of EIAV MA.