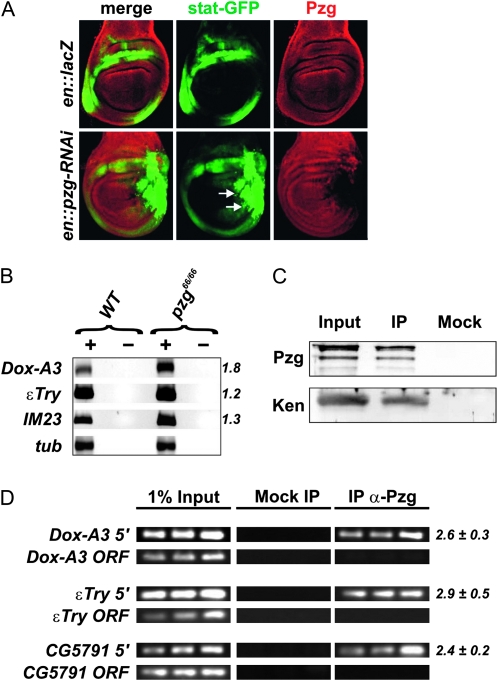
Figure 6.—
Pzg antagonizes JAK/STAT-signaling activity. (A) pzg-RNAi induction in the posterior compartment of the wing disk increases the expression of a STAT92E-GFP reporter (arrows). Control in top row: en-Gal4::UAS-lacZ/STAT92E-GFP; bottom row: en-Gal4::UAS-pzg-RNAi/STAT92E-GFP. Wing disks were stained with anti-Pzg antibodies (red). Posterior is to the right, dorsal is up. (B) The transcript levels of STAT responsive genes Dox-A3, εTry, and IM23 were determined by semiquantitative RT–PCR. The levels were normalized to β-tubulin (tub). As a control, reactions were performed with (+) and without (−) reverse transcriptase. (C) Pzg can be detected in a complex with the repressor Ken in vivo. Proteins immunoprecipitated (IP) from larval protein extracts by anti-Pzg antibodies were probed to detect Ken proteins. The input lane contained 25% of the larval extract used for the IP. A mock control was performed with preimmune serum to demonstrate specificity. (D) XChIP analyses showing that Pzg can be localized at the promoter regions of Dox-A3, εTry, and CG579, but it is absent from the respective coding regions. Primer sets span regulatory regions that contain predicted Ken and NURF binding sites. Samples of the 31st, 33rd, and 35th PCR amplification cycles are shown. Relative enrichment was estimated for the 33rd PCR cycle sample from the ratio between Pzg IP and mock signals. The mean values and standard deviations of at least three independent experiments were calculated. PCR amplification of regions within the ORF of Dox−A3, εTry, and CG5791 were performed as an unrelated control.