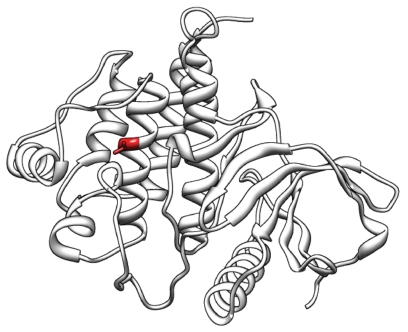
Figure 2.
Protein structure of ZAP70 and structural variations by nsSNPs. We predicted the structural changes in ZAP70 caused by nsSNPs through structural modeling and stability analysis because mutations can cause selective T-cell defects [46]. In order to predict the structure of ZAP70, we used PDB entries 1M61 and 1U59 as the structural templates, which have 100% and 98.9% sequence identity, respectively, which were found to be suitable structural templates. We found 19 nsSNPs from the dbSNP database that cause nonsense or missense changes in ZAP70. From these nsSNPs, 10 nsSNPs were found to lead to unstable structural changes by using CUPSAT. In particular, Val527Gly had the lowest unstable energy value (Predicted ΔΔG: −4.09). The red region shows the location of Val527Gly in the middle of the alpha-helix structure.