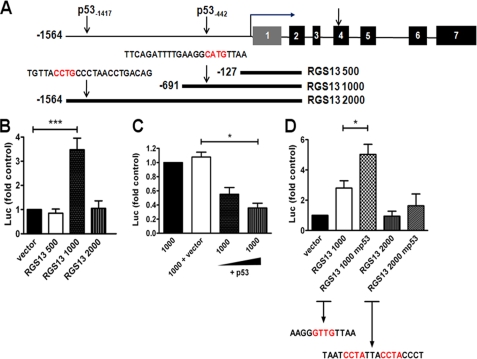
FIGURE 2.
Characterization of the RGS13 promoter. A, the locations of two p53 REs in the 5′-flanking region of RGS13 and a schematic representation of luciferase reporter constructs are shown. PCR products encompassing the denoted regions were amplified from genomic DNA and subcloned into a promoterless luciferase reporter vector (pGL3-Basic). Numbers are relative to the experimentally derived TSS (bent arrow). B and C, RGS13 promoter activity. Luciferase reporter constructs were transfected into LAD2 cells together with a plasmid encoding Renilla luciferase. Luciferase (Luc) activity in cell extracts was measured after 24 h. Data are means ± S.E. of activity relative to cells expressing empty control vector (or RGS13 1000 in C) in three or more independent experiments. *, p = 0.02; ***, p < 0.0001 (one-way ANOVA). D, relative luciferase activity of WT or mutant (mp53) constructs lacking core p53-binding motifs. The mutated sequence is denoted in red. *, p < 0.05 (one-way ANOVA).