FIGURE 1.
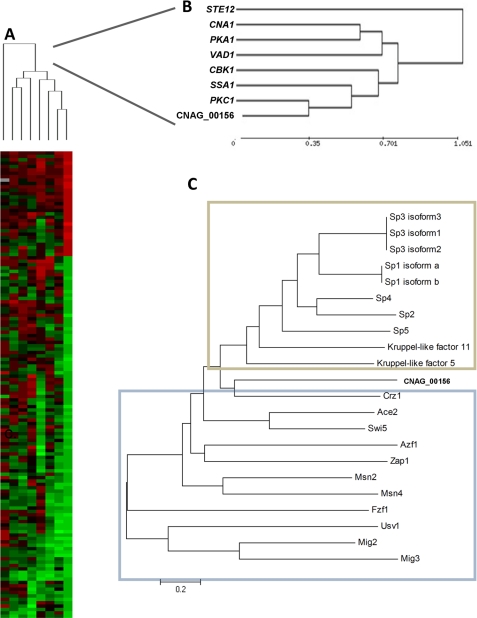
Transcriptional and protein sequence comparison of a C-terminal zinc finger (CNAG_00156). A, heat map display of a comparative microarray experiment of C. neoformans strains Cn sp1Δ (cnag_00156Δ), pkc1Δ, pka1Δ, cna1Δ, cbk1Δ, ssa1Δ, ste12Δ, and vad1Δ. Strains were grown overnight to mid-log phase and induced in 0% glucose medium for 1 h before RNA extraction. WT (H99) induced under the same conditions was used as reference for each mutant. A cluster analysis of genes significantly altered in mutant strains with p < 0.0001 is presented. B, close-up view of the cluster analysis presented in A. C, neighbor-joining phylogenic analysis of the Cn Sp1 (Cnag_00156) with S. cerevisiae (blue frame) and H. sapiens (tan frame) C-terminal zinc finger transcription factors.