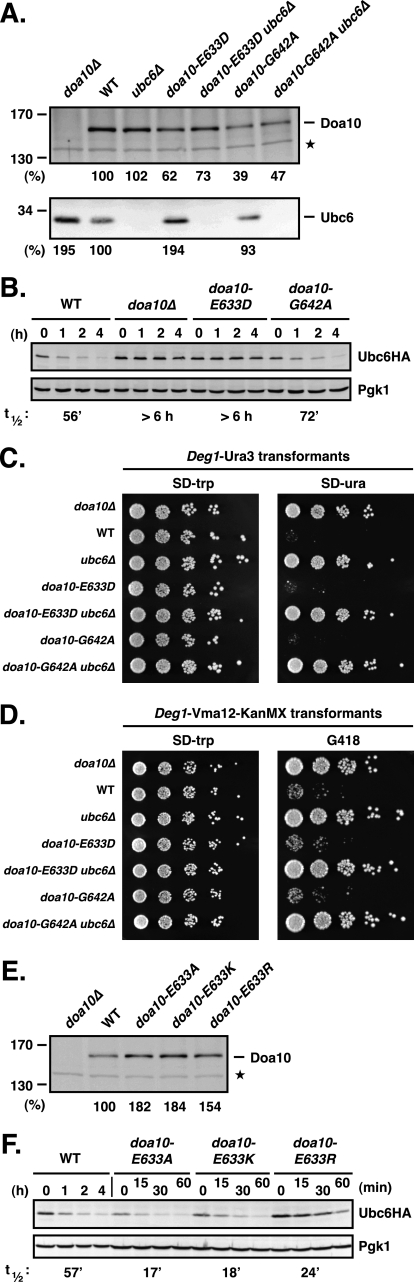
FIGURE 5.
E633D mutation in TM5 of Doa10 strongly decreases degradation of Ubc6 but has no detectable effect on other substrates. A, comparison of Doa10, doa10-E633D, and doa10-G642A protein levels in the presence and absence of Ubc6 and the corresponding Ubc6 steady state levels. Immunoblot analysis and quantitation was done as in Fig. 3B. Doa10 and Ubc6 levels in WT were set to 100%. Asterisk, a cross-reacting yeast protein used to compare protein loading. B, conservative doa10-E633D substitution leads to stabilization of Ubc6HA. Cycloheximide chase/anti-HA immunoblot analysis was done as in Fig. 2B. C, degradation of the soluble substrate Deg1-Ura3 in doa10-E633D cells (and doa10-G642A cells) inferred from growth assays. Degradation of Deg1-Ura3 is dependent on the presence of Ubc6. Serial dilutions of the indicated strains that had been transformed with a TRP1-marked Deg1-Ura3 expression plasmid were spotted onto minimal plates lacking tryptophan (SD−trp) or uracil (SD−ura) and incubated for 2.5 days at 30 °C. D, degradation of the membrane substrate Deg1-Vma12-KanMX in doa10-E633D cells (as well as doa10-G642A cells) inferred from growth assays. Degradation of Deg1-Vma12-KanMX is dependent on the presence of Ubc6. Serial dilutions of the indicated strains that had been transformed with a TRP1-marked Deg1-Vma12-KanMX6 expression plasmid were spotted onto a minimal plate lacking tryptophan (SD−trp) or on a YPD plate supplemented with 300 μg/ml G418 and incubated for 2 days at 30 °C. E, steady state levels of Doa10 and different doa10-E633 substitutions (Ala, Lys, or Arg). Immunoblot analysis and quantitation was done as in Fig. 3B. F, doa10-E633A, doa10-E633K, or doa10-E633R mutations all accelerate Ubc6HA degradation. Cycloheximide chase/anti-HA immunoblot analysis was done as in Fig. 2B.