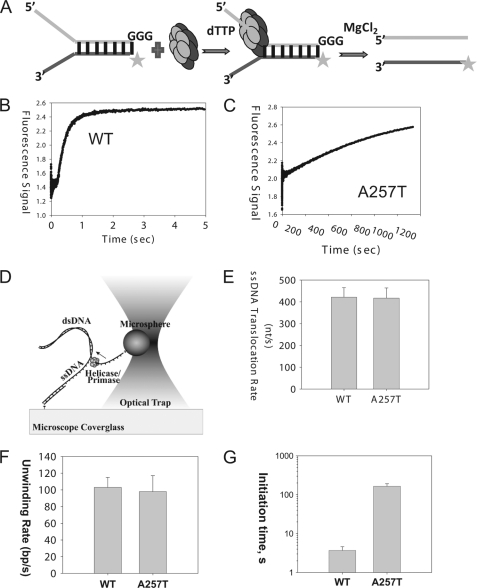
FIGURE 2.
Helicase activities. A, scheme showing the ensemble unwinding assay. The ring-shaped hexameric helicase loads on the 5′-overhang and moves from the 5′-end to the 3′-end to unwind the fluorescein-labeled dsDNA fork substrate. T7 gp4 WT or A257T (100 nm) was preincubated with the fluorescent ds30 (23%) substrate (10 nm) and dTTP (2 mm) in one syringe of the stopped-flow instrument, and the unwinding reaction was initiated by addition of MgCl2 from a second syringe at 25 °C. B, unwinding kinetics of WT gp4. The lag kinetics was fit to the incomplete gamma function (29) to obtain a rate of ∼67 bp/s for the WT gp4. C, fluorescence signal resulting from unwinding of dsDNA into ssDNA by A257T plotted against time. The kinetics fit to a single exponential rate constant of 1.3 × 10−3 s−1. D, scheme of the single molecule optical trap setup (not to scale). A helicase/primase molecule separates the dsDNA, while a constant force is maintained on the DNA fork junction by adjusting the position of the glass coverslip relative to the optically trapped microsphere. The displacement of the trap relative the coverslip is utilized to determine the amount of DNA unwound (and subsequently the unwinding rate). E, comparison of ssDNA translocation rate of WT gp4 and A257T. Experiments were carried out at 0.3 nm WT and 30 nm A257T, 2 mm dTTP, 5 mm MgCl2. F, comparison of the unwinding rate WT gp4 and A257T. G, comparison of the initiation times of DNA unwinding by WT gp4 and A257T. The initiation times were experimentally determined at 5 nm hexamer, 2 mm dTTP, 5 mm MgCl2. The data represent average from >20 traces.