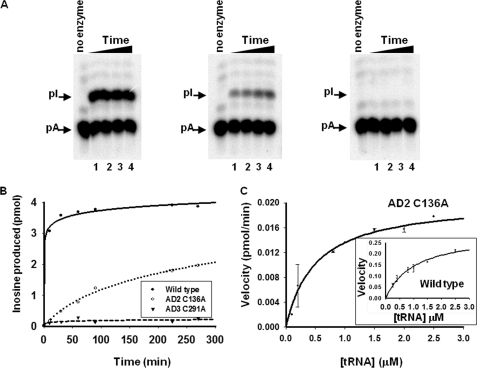
FIGURE 2.
Amino acid substitution of one of the key zinc-coordinating residue in TbADAT2 yields an active enzyme. A, a site-specifically 32P-labeled (at A34) tRNA substrate was incubated with constant concentrations of wild type and mutant enzyme for varying time. The reaction products were then digested to 5′-nucleotide monophosphates and separated by thin layer chromatography. The arrows indicate the positions of 5′-adenosine monophosphate (pA) and the product 5′-inosine monophosphate (pI). Nonradioactive nucleotides were used as markers (not shown). In all panels, the no enzyme lane refers to a negative control in which tRNA was incubated with buffer in the absence of enzyme; this also served as a control for background during quantitation. The samples were incubated with wild type or mutant enzymes for 60, 90, 120, and 180 min (lanes 1–4, respectively) (panels from left to right, wild type, AD C136A, and AD3 C291A). B, the left panel shows a graph of a time course of similar reactions as above but for extended periods of time. AD2 C136A denotes a substitution of one of the zinc-coordinating cysteines in TbADAT2 expressed in the context of a heterodimer with wild type TbADAT3. AD3 C291A denotes the analogous mutant but in TbADAT3 in the context of wild type TbADAT2. C, steady-state kinetic analysis of the TbADAT2 C136A mutant by incubation with increasing saturating concentrations of tRNA substrate. The reaction velocity (pmol of inosine formed per minute) was plotted against tRNA substrate concentration. This curve was fit to the Michaelis-Menten equation using SigmaPlot software. The inset shows a similar experiment but with the recombinant wild type enzyme. Each plot represents at least three independent experiments.