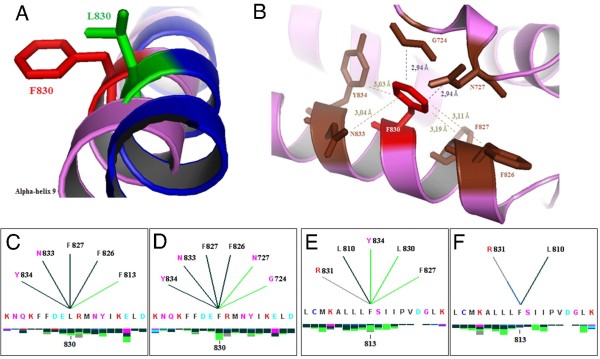
Figure 3.
Modeling for normal and mutant AR proteins. (A) Comparison of normal and mutant human AR protein models at residue 830; normal leucine is denoted in green and phenylalanine in red. (B) Distances in Ångström (Å) for contacts for the F830 mutant residue estimated using PyMOL software: hydrogen bonds are shown in brown and hydrophobic interactions in purple. (C - F) Internal contacts provided by the analysis with BlueStar STING software. The native residue L830 (C) forms energetic hydrogen bonds with F826, F827, N833 and Y834 and a hydrophobic interaction with F813, whereas the mutant residue F830 (D) suppresses the interaction with F813 and introduces two additional hydrophobic interactions with G724 and N727. Effects upon internal contacts for F813 residue: in the normal protein, the F813 residue presents hydrophobic interaction with F827 and Y834 (E), whereas in the mutant protein those interactions are lost (F).