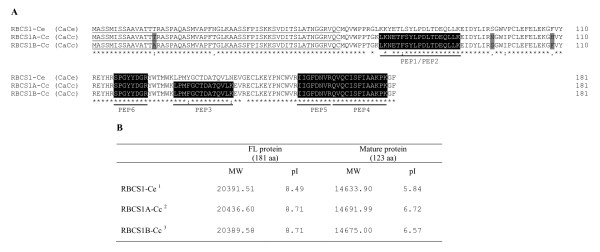
Figure 3.
Sequence alignment and characteristics of the coffee RBCS1 proteins. (A): The amino acids corresponding to the chloroplastic transit peptide [1 to 58] are underlined. Identical amino acids are indicated by stars, conservative substitutions are indicated by two vertically stacked dots and semi-conservative substitutions are indicated by single dots. The RBCS1-Ce (CaCe) isoform from C. eugenioides corresponded to the proteins with the GenBank accession numbers CAD11990 and CAD11991 translated from the CaRBCS1 cDNA [GenBank:AJ419826] and gene [GenBank:AJ419827], respectively. The RBCS1A-Cc (CaCc) protein from the CcRBCS1 cDNA (FR728242) and gene (FR772689) sequences of C. canephora (this study) was strictly identical to the protein deduced from the SGN-U617577 unigene. The RBCS1B-Cc (CaCc) protein was deduced from the SGN-U607190 unigene. Divergent amino acids between RBCS1-Ce (CaCe) and RBCS1A-Cc (CaCc) proteins are boxed in grey, and those confirmed by mass spectrometry analysis (Table 6) are boxed in black. (B) The RBCS1-Ce (CaCe) protein deduced from the CaRBCS1 cDNA and gene sequences was identical to the protein deduced from the SGN-U607188 unigene (1). The RBCS1A-Cc protein was deduced from the RBCS1-Cc (identical to SGN-U6175772) cDNA and gene sequences from C. canephora (this study). The RBCS1B-Cc protein was deduced from the SGN-U607190 (3) nucleic acid sequence. Molecular weights (MW in Daltons), amino acids (aa) and isoelectric points (pI) are indicated for full-length (FL) and mature (without the chloroplast transit peptide) RBCS1 proteins. SGN sequences were obtained from the Sol Genomics Network http://solgenomics.net/content/coffee.pl.