Table 2. GWAS results for all SNPs with 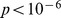 in the 23andMe cohort.
in the 23andMe cohort.
| SNP | Chr | Position | Region | Alleles | MAF | Cohort | OR |
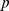
|
| rs34637584 | 12 | 39020469 | LRRK2 | G/A | 0.002 | 23andMe | 9.615 (6.43–14.37) |
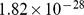
|
| IPDGC | – | – | ||||||
| i4000416 | 1 | 153472258 | GBA | T/C | 0.005 | 23andMe | 4.048 (3.08–5.32) |
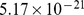
|
| IPDGC | – | – | ||||||
| rs356220 | 4 | 90860363 | SNCA | C/T | 0.375 | 23andMe | 1.285 (1.22–1.36) |
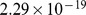
|
| IPDGC | – | – | ||||||
| rs12185268 | 17 | 41279463 | MAPT | A/G | 0.211 | 23andMe | 0.769 (0.72–0.82) |
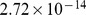
|
| IPDGC | – | – | ||||||
| rs10513789 | 3 | 184242767 | MCCC1/LAMP3 | T/G | 0.201 | 23andMe | 0.803 (0.75–0.86) |
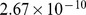
|
| IPDGC | 0.873 (0.83–0.92) |
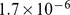
|
||||||
| rs6812193 | 4 | 77418010 | SCARB2 | C/T | 0.365 | 23andMe | 0.839 (0.79–0.89) |
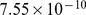
|
| IPDGC | 0.90 (0.86–0.94) |
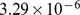
|
||||||
| rs6599389 | 4 | 929113 | GAK | G/A | 0.075 | 23andMe | 1.311 (1.19–1.44) |
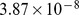
|
| IPDGC | – | – | ||||||
| rs11868035 | 17 | 17655826 | SREBF1/RAI1 | G/A | 0.309 | 23andMe | 0.851 (0.80–0.90) |
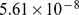
|
| IPDGC | 0.95 (0.91–0.996) | 0.033 | ||||||
| rs823156 | 1 | 204031263 | SLC41A1 | A/G | 0.183 | 23andMe | 0.827 (0.77–0.89) |
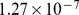
|
| IPDGC | – | – | ||||||
| rs4130047 | 18 | 38932233 | RIT2/SYT4 | T/C | 0.313 | 23andMe | 1.161 (1.10–1.23) |
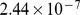
|
| IPDGC | 1.077 (1.03–1.13) | 0.0014 | ||||||
| rs2823357 | 21 | 15836776 | USP25 | G/A | 0.376 | 23andMe | 1.149 (1.09–1.21) |
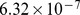
|
| IPDGC | 0.971 (0.93–1.02) | 0.187 |
All genomic positions are given with respect to NCBI build 36.3. Alleles are listed as major/minor and are specified for the forward strand. Odds ratios per copy of the minor allele and 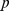 -values are provided for the 23andMe cohort and, where requested, the IPDGC replication set. Minor allele frequencies are provided for the 23andMe cohort.
-values are provided for the 23andMe cohort and, where requested, the IPDGC replication set. Minor allele frequencies are provided for the 23andMe cohort.
