Table 5. Internal and external cross-validation experiments using sparse logistic regression.
| Internal Validation | External Validation | |||||
| Signif. Threshold | Avg SNPs | Avg Regions | AUC | SNPs | Regions | AUC |
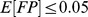
|
9.0 | 6.6 |
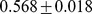
|
11 | 9 |
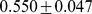
|
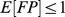
|
18.4 | 15.4 |
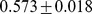
|
22 | 19 |
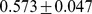
|
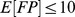
|
41.6 | 35.0 |
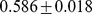
|
60 | 51 |
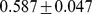
|
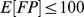
|
156.0 | 138.8 |
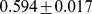
|
220 | 195 |
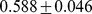
|
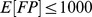
|
698.4 | 639.2 |
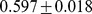
|
803 | 727 |
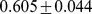
|
The internal five-fold cross-validation experiment was performed using only the 23andMe cohort. The external cross-validation experiment was performed by training on the 23andMe cohort and testing on the NINDS cohort. “SNPs” denotes the number of SNPs included in the fitted model. “Regions” denotes the number of distinct LD blocks represented by the SNPs in the fitted model. Each AUC value represents a covariate-adjusted AUC. For the internal validation experiment, average values are provided for SNPs and Regions, providing an average over all five cross-validation folds, and AUCs were computed by pooling predictions over the five cross-validation folds. For each row of the table, the sparsity inducing prior was chosen to achieve the approximate upper bound on the expected false positive rate indicated in the first column; here, 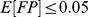 corresponds to a model containing only genome-wide significant associations, whereas
corresponds to a model containing only genome-wide significant associations, whereas 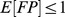 corresponds to suggestive associations. In each of the internal and external validation experiments, models with AUCs in bold are significantly better than non-bold models (see Table S3).
corresponds to suggestive associations. In each of the internal and external validation experiments, models with AUCs in bold are significantly better than non-bold models (see Table S3).
