Abstract
The highly conserved ANP32 proteins are proposed to function in a broad array of physiological activities through molecular mechanisms as diverse as phosphatase inhibition, chromatin regulation, caspase activation, and intracellular transport. On the basis of previous analyses of mice bearing targeted mutations of Anp32a or Anp32e, there has been speculation that all ANP32 proteins play redundant roles and are dispensable for normal development. However, more recent work has suggested that ANP32B may in fact have functions that are not shared by other ANP32 family members. Here we report that ANP32B expression is associated with a poor prognosis in human breast cancer, consistent with the increased levels of Anp32b mRNA present in proliferating wild-type (WT) murine embryonic fibroblasts and stimulated WT B and T lymphocytes. Moreover, we show that, contrary to previous assumptions, Anp32b is very important for murine embryogenesis. In a mixed genetic background, ANP32B-deficient mice displayed a partially penetrant perinatal lethality that became fully penetrant in a pure C57BL/6 background. Surviving ANP32B-deficient mice showed reduced viability due to variable defects in various organ systems. Study of compound mutants lacking ANP32A, ANP32B, and/or ANP32E revealed previously hidden roles for ANP32A in mouse development that became apparent only in the complete absence of ANP32B. Our data demonstrate a hierarchy of importance for the mammalian Anp32 genes, with Anp32b being the most critical for normal development.
Keywords: cell growth, mouse embryogenesis, premature aging
ANP32B (a.k.a. APRIL, PAL31, PHAP1b) is a mammalian member of the highly conserved acidic nuclear phosphoprotein 32kDa (ANP32) family of gene products (1). These metazoan-specific factors are characterized by the presence of an amino-terminal leucine-rich repeat domain and a carboxyl-terminal region that is highly enriched in acidic amino acid residues (2). These features are found in ANP32 proteins from mapmodulin, the single representative in Drosophila, and in the three vertebrate family members identified ANP32A, ANP32B, and ANP32E (1, 3).
The ANP32 proteins have been implicated in a broad array of physiological processes, including cell differentiation (4–7), apoptotic cell death (8–14), and cell proliferation (15–17). Diverse mechanisms have been postulated for how these proteins perform their function(s). Some studies indicate that ANP32 proteins may directly control enzymatic activities, such as via inhibition of protein phosphatase 2A (PP2A) (18–20) or activation of caspases (10, 11, 13, 14, 21). Other studies suggest that they may regulate intracellular transport at nuclear pores or microtubules (22–25). Several studies present evidence that nuclear ANP32 proteins may influence transcription either through the inhibitor of acetyl transferases complex (26–29) or by direct effects on transcription factors (30, 31). Most of the above reports have focused on ANP32A, the founding member of the ANP32 family, but none of these studies has specifically excluded a particular ANP32 protein as contributing to the activities examined.
More recent work has demonstrated functions that are exclusive to the ANP32B protein, at least in humans. First, ANP32B, but not ANP32A, controls the expression of the dendritic cell maturation factor CD83 by regulating the transport of its mRNA to the cytoplasm (22). Second, ANP32B modulates the activity of the transcription factor Kruppel-like factor 5 (KLF5), whereas ANP32A cannot (32). Finally, ANP32B is a caspase substrate, whereas ANP32A is not (33).
Reported loss-of-function mutants for ANP32 family members include two independently targeted ANP32A-deficient mice (34, 35), an ANP32E-deficient mouse (35), and a presumptive null mutant of mapmodulin in Drosophila (36). All of these mutants were viable and fertile with no obvious abnormalities. Here, we report on the phenotype of Anp32b-deficient (Anp32b−/−) mice and, using compound mutants, show that ANP32 family members are not completely redundant in mammals. We demonstrate that ANP32B is the most important ANP32 family member for mammalian development and likely plays a complex role in sustaining viability that has yet to be completely defined. In addition, we provide evidence that the level of ANP32B mRNA expression in human breast cancers may serve as a prognostic marker.
Results
ANP32B as a Potential Prognostic Marker in Human Cancer.
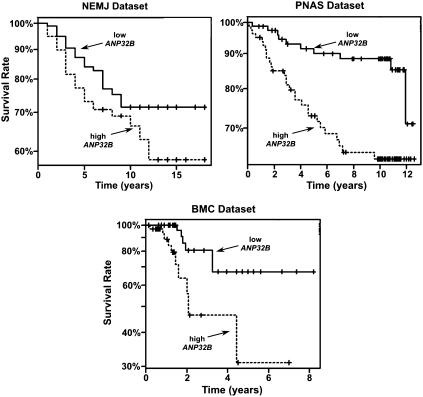
Reports that ANP32B expression was related to cell proliferation (15, 17) prompted us to investigate whether ANP32B expression was altered in human tumor samples. We examined the relationship between ANP32B mRNA expression and breast cancer patient prognosis using information from three available studies, namely the NEJM (New England Medical Journal) (37), the BMC (BMC Genomics) (38), and the PNAS datasets (39). When we compared the survival of these three groups of patients among all three datasets, we found that patients whose tumors showed the highest ANP32B mRNA levels had significantly shorter survival times using the Cox proportional hazard model (P < 10−4). To display this effect, we stratified the patients in these datasets into three major groups: those with tumors expressing high levels of ANP32B mRNA (highest 33.3% or tertile), those with tumors with medium ANP32B mRNA levels (middle tertile), and those with tumors with low ANP32B mRNA levels (lowest tertile). Here, the survival effect of the highest and lowest ANP32B-expressing tumors is clearly demonstrated (Fig. 1; middle tertile not shown). These results imply that elevated ANP32B expression in a tumor may be a marker of poor patient prognosis.
Fig. 1.
ANP32B mRNA expression is a marker for aggressive breast cancer. Survival analyses of patients with tumors expressing the highest levels of ANP32B mRNA (highest tertile: dashed line), and patients with tumors expressing the lowest levels of ANP32B mRNA (lowest tertile: solid line). The middle tertile is not shown. Values were acquired from three separate datasets: NEMJ (37), PNAS (39), and BMC (38).
Anp32b mRNA Levels Are Linked to Cell Proliferation in Mice.
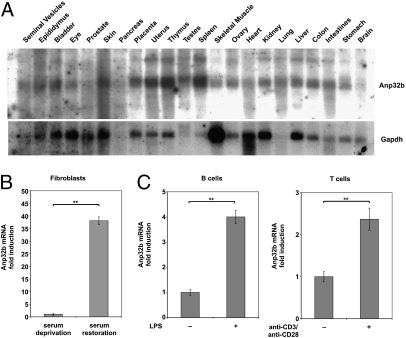
The less favorable prognosis of breast cancer patients with high ANP32B-expressing tumors suggested that the elevated ANP32B in these samples might be linked to more aggressive tumor cell proliferation. To investigate this hypothesis, we performed Northern blot analysis of a wide range of tissues from adult wild-type (WT) mice. We found that Anp32b expression was low (relative to Gapdh) in tissues where less cell proliferation usually occurs, i.e., in brain tissue (Fig. 2A). In contrast, tissues that generally have high cell proliferation rates, i.e., spleen and thymus, showed higher relative levels of Anp32b mRNA expression. We confirmed this pattern at the mRNA level by quantitating Anp32b expression (relative to Tbp) using quantitative RT-PCR (Fig. S1A). Again, proliferative tissues such as the spleen and thymus showed elevated levels of Anp32b mRNA, whereas the forebrain and hindbrain exhibited only low Anp32b expression. These data further suggest a link between Anp32b level and rate of cell proliferation.
Fig. 2.
Elevated Anp32b mRNA expression correlates with increased cell proliferation. (A) Correlation with highly proliferative tissues. A commercial mouse tissue blot was examined by Northern blotting to detect Anp32b mRNA. Gapdh: relative expression control. Anp32b mRNA is decreased relative to Gapdh in brain and muscle. (B) Correlation with nutrient-induced proliferation. WT MEFs were deprived of serum for 72 h and then resupplied with medium containing 10% FBS for 24 h. Anp32b mRNA levels were determined by qPCR relative to β-actin. Data shown are the mean ± SD of two independent WT MEF cultures. **P < 0.01. (C) Correlation with lymphocyte stimulation. (Left) Purified B cells from WT mice were treated with 1 μg/mL LPS for 24 h. (Right) Purified T cells from WT mice were stimulated with plate-bound anti-CD3 (2 μg/mL) plus anti-CD28 (0.2 μg/mL) for 36 h. In both cases, Anp32b mRNA levels were determined by qPCR relative to 18S rRNA. Results shown are the mean ± SD of triplicates and are expressed as fold increase over levels in untreated controls. Results are representative of two trials. **P < 0.01.
To test whether proliferative stimuli could induce Anp32b expression, we subjected WT primary mouse embryo fibroblasts (MEFs) to serum deprivation to induce cell cycle arrest and then stimulated their proliferation by restoring standard serum levels to the culture. Under conditions of serum deprivation, Anp32b mRNA was barely detectable in WT MEFs (Fig. 2B). However, upon the addition of serum and the resumption of proliferation, Anp32b mRNA was significantly increased (P < 0.01) by more than 38-fold relative to control Actb. Under identical conditions, the induction of mRNA encoding Myc, a well-documented early proliferation marker, achieved a 4.8-fold increase (Fig. S1B). Similarly, WT B cells stimulated with LPS, and WT T cells activated with anti-CD3 plus anti-CD28 antibodies (activators of the T-cell receptor complex and T-cell costimulatory complex, respectively), showed significant elevation (P < 0.01) of Anp32b mRNA levels (Fig. 2C). These results indicate that Anp32b is up-regulated under conditions that favor cell proliferation.
Gene Targeting of Anp32b in Mice.
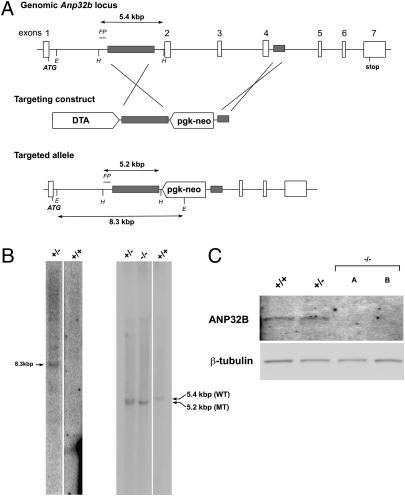
To investigate the physiological functions of ANP32B in vivo, we generated mice in which the Anp32b gene was disrupted by replacing the genomic region containing exons 2, 3, and 4 with a pgk-neo expression cassette (Fig. 3A). This mutation results in an mRNA in which the last 17 codons of exon 1 are linked to an out-of-frame exon 5. Southern blotting of the genomic DNA of E14K ES cell clones demonstrated a single appropriate insertion of the pgk-neo sequence (Fig. 3B, Left). Anp32b+/− mice (mixed 129:C57BL/6 background) were then generated and intercrossed to create Anp32b−/− progeny. Genomic deletion was confirmed in primary MEFs derived from Anp32b−/− embryos using a flanking genomic probe (Fig. 3B, Right). Immunoblotting of extracts of Anp32b−/− MEFs demonstrated that there was no detectable Anp32b protein in nullizygous mutants (Fig. 3C).
Fig. 3.
Generation and validation of ANP32B-deficient mice. (A) Targeting of the murine Anp32b gene. (Top) Murine genomic Anp32b locus showing exons and regions of homology (gray). FP: flanking probe detecting intron 1. H: HindIII; E: EagI sites. The translation start site (ATG) and stop sites are shown. (Middle) Targeting construct that replaced exons 2–4 with a pgk-neo cassette. DTA: diphtheria toxin A sequence for negative selection. (Bottom) Targeted Anp32b allele. Diagnostic HindIII fragments for the WT Anp32b allele (5.4 kb) and the targeted Anp32b allele (5.2 kb), as well as the diagnostic EagI fragment for neo insertion (8.3 kb) are shown. (B) Confirmation of Anp32b gene deletion. (Left) DNA from Anp32b+/− and Anp32b+/+ ES cells was Southern-blotted with a neo probe to confirm a single vector insertion. (Right) DNA from MEFs derived from Anp32b+/−, Anp32b−/−, or Anp32b+/+ mice was Southern-blotted using the flanking probe. WT HindIII band = 5.4 kb; mutant (MT) HindIII band = 5.2 kb. (C) Loss of ANP32B protein. Anp32b+/+, Anp32b+/− and Anp32b −/− MEFs were immunoblotted to detect ANP32B protein. A and B are MEFs from two different Anp32b−/− littermate embryos. β-Tubulin: loading control. Results shown in B and C are representative of three trials.
ANP32B Protects Against Perinatal Lethality.
The intercrossing of our mixed-bred 129:C57BL/6 Anp32b+/− mice to generate Anp32b−/− mutants revealed a defect in mutant mouse viability. At the time of weaning (∼21 d of age), Anp32b−/− mice were present at a far lower frequency (7%) than the 25% expected according to the standard Mendelian distribution (P < 0.01; Table 1). To exclude the possibility that the observed defect was due to a linked mutation present in the ES cell clones, we backcrossed the Anp32b mutant allele into the C57BL/6 background for six generations. The penetrance of the survival defect was increased in the pure C57BL/6 background (P < 0.01; Table 2), strongly suggesting that the defect in viability is not attributable to a linked gene mutation.
Table 1.
Reduced survival rate of Anp32b-deficient mice
| Anp32b+/+ | Anp32b+/− | Anp32b−/− | |
| Expected | 39 | 78 | 39 |
| Observed* | 57 | 87 | 11 |
Anp32b+/− mice of mixed 129ola:C57BL/6 background were intercrossed and pups were genotyped at time of weaning (between postnatal 19 and 22 d). “Expected” indicates the distribution for the indicated total number of mice under conditions of normal Mendelian segregation. “Observed” indicates the actual numbers of mice of the indicated genotypes obtained.
*P < 0.01 by χ2 analysis.
Table 2.
Genetic background influence survival rate of Anp32b-deficient mice
| Anp32b+/+ | Anp32b+/− | Anp32b−/− | |
| Expected | 36 | 72 | 36 |
| Observed* | 56 | 87 | 1 |
Anp32b+/− mice from congenic C57BL/6 background were intercrossed and pups were genotyped at time of weaning (between postnatal 19 and 22 d). “Expected” indicates the distribution for the indicated total number of mice under conditions of normal Mendelian segregation. “Observed” indicates the actual numbers of mice of the indicated genotypes obtained.
*P < 0.01 by χ2 analysis.
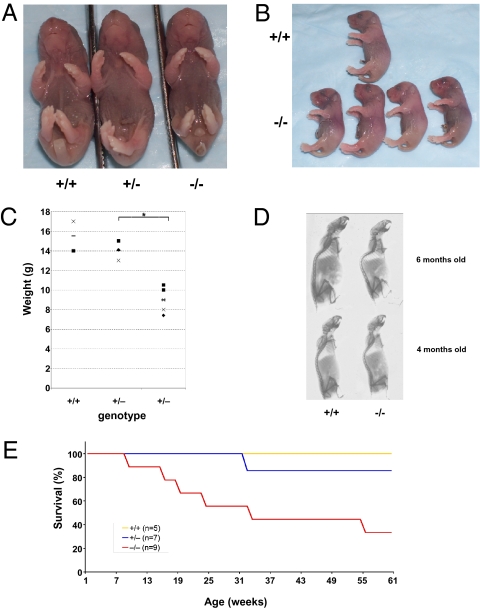
To define precisely when the majority of Anp32b−/− mice died, we monitored litters from the time of first detection (12 h postnatal) until weaning. However, virtually all Anp32b−/− mice that were evident at first detection survived to adulthood. This observation indicated that the developmental problem(s) killing most Anp32b−/− mice occurred either during embryogenesis or before normal litter detection. In the course of generating our primary MEFs, we noted that Anp32b−/− embryos were present at approximately Mendelian rates on embryonic day 14.5 (E14.5). We therefore chose to follow the fate of Anp32b−/− embryos from late embryogenesis until early time points after birth. Among litters examined immediately after birth, some Anp32b−/− pups never initiated breathing, whereas others were able to nurse, at least initially. Timed pregnancy studies revealed that E17.5 Anp32b−/− embryos were present at approximately Mendelian frequency (Table 3). No defect detectable by gross inspection was evident in any E17.5 Anp32b−/− embryos of the mixed background (Fig. 4 A and B). Examination of Anp32b−/− embryos of the pure C57BL/6 background showed that these animals also remained viable until at least E17.5. Taken together, our data suggest that the defect(s) killing Anp32b−/− mice occur perinatally, i.e., at or very near the time of birth.
Table 3.
Normal survival rate of Anp32b-deficient embryos at E17.5
| Anp32b+/+ | Anp32b+/− | Anp32b−/− | |
| Expected | 6 | 11 | 6 |
| Observed* | 8 | 8 | 6 |
Anp32b+/− mice were intercrossed and embryos were genotyped at time E17.5. “Expected” indicates the distribution for the indicated total number of embryos under conditions of normal Mendelian segregation. “Observed” indicates the actual numbers of embryos of the indicated genotypes obtained.
*No significant differences were found.
Fig. 4.
Phenotypes of ANP32B-deficient mice. (A and B) Normal gross appearance. Embryos of the indicated Anp32b genotypes were collected at E17.5 and subjected to gross examination. (A) Rostral view. (B) Left side view. Tail fragments were removed for genotyping purposes. No obvious abnormalities were seen. Embryos shown were derived from two different litters and are representative of a total of 10 examined per genotype. (C) Reduced body weight. Anp32b−/− mice were weighed alongside their Anp32b+/+ or Anp32b+/− littermates at 3 wk of age. Results shown are values for individual mice. Horizontal bars are mean values. *P < 0.05. (D) Kyphosis. X-ray analysis was performed on littermate pairs of female Anp32b+/+ and Anp32b−/− mice at 4 and 6 mo of age. Results shown are representative of over 10 mice examined per genotype per age. (E) Decreased life span. Survival curves of Anp32b+/+, Anp32b+/− and Anp32b−/− littermates that lived past weaning age are shown. The reduced survival of Anp32b−/− mice is statistically significant, as determined by log-rank analysis (P < 0.01).
Rigorous pathological examinations of eight Anp32b−/− E17.5 embryos as well as three postnatal day 1 (P1) pups of both the mixed 129:C57BL/6 and the pure C57BL/6 backgrounds revealed no obvious gross abnormalities in a majority of embryos examined. Some Anp32b−/− E17.5 embryos did show subtle and sporadic craniofacial abnormalities, including overlarge ventricles in the brain, expanded inner ear cavities, and palate closure defects. The heart and lungs were not obviously affected. Interestingly, large hematomas in the liver, aortic arch, or umbilical artery were observed in three of eight E17.5 embryos and in one of three P1 pups of the mixed mutant background. However, no one defect appeared to be responsible for the perinatal lethality of all Anp32b−/− mice.
Surviving Anp32b−/− Mice Show Reduced Size and Decreased Life Span.
To investigate the effects of ANP32B deficiency in adult mice, we monitored our surviving mixed-bred Anp32b−/− mice and their littermates from time of weaning until age approximately 1 y. At time of weaning, surviving Anp32b−/− female mice weighed, on average, 42% less than WT females and 35% less than their female Anp32b+/− littermates (P < 0.05; Fig. 4C). A similar trend was seen for males, although the differences did not reach statistical significance in this analysis (Fig. S2). When the data for male and female sex-matched littermates were combined, a statistically significant reduction in body weight was apparent when Anp32b−/− and Anp32b+/− littermates were compared (P < 0.01), as well as when Anp32b−/− and Anp32b+/+ littermates were compared (P < 0.05).
Surviving Anp32b−/− mice were not robust and showed signs of premature aging. Many Anp32b−/− mutants exhibited early onset kyphosis in which the curvature of the upper spine was evident at 4 mo of age and severe by 6 mo (Fig. 4D). Increased hepatocyte polyploidy, another hallmark of premature aging, was also present in the aged Anp32b−/− mice. However, premature alopecia, which is a common aspect of early aging, was not seen in the kyphotic mice. Consistent with these phenotypic features, Anp32b−/− mice had a reduced life span (P < 0.01; Fig. 4E), but no consistent pathology could be identified as the cause of their premature death. Two cases of megacolon and two cases of hydroureter causing severe morbidity were observed, but the other five aged Anp32b−/− mice died without identifiable cause.
Normal Cellular Apoptosis and Proliferation in the Absence of ANP32B.
We next turned to in vitro studies in an attempt to identify the cause of the viability defects in Anp32b−/− mice. Because Anp32b has been implicated in the regulation of caspase activity (10, 33), we first investigated the response of Anp32b−/− thymocytes and transformed MEFs to a broad array of intrinsic and extrinsic apoptotic stimuli. However, no statistically significant differences between Anp32b+/+ and Anp32b−/− cells of either type were observed (Fig. S3). Similarly, prompted by the conclusions of previous reports (15, 17), we investigated whether loss of ANP32B affected cell proliferation, but, again, no defects in in vitro proliferation were found for Anp32b−/− primary MEFs, thymocytes, or splenocytes (Fig. S4).
ANP32B Masks a Role for ANP32A in Essential Development.
We have previously reported that neither the ANP32A-deficient nor the ANP32E-deficient mouse has any obvious abnormal phenotype (35). To determine whether deficiency for another Anp32 family member might expose the cause of the viability defect of our Anp32b−/− mice, we generated Anp32b+/−;Anp32a+/− and Anp32b+/−;Anp32e+/− double-mutant strains and intercrossed them. As shown in Table 4, heterozygosity for Anp32a further compromised development in the Anp32b−/− background (P < 0.01). Indeed, no Anp32b−/−;Anp32a+/− or Anp32b−/−;Anp32a−/− mouse survived to weaning. Thus, contrary to prevailing belief, ANP32A does have a subtle role in murine embryogenesis that is revealed only in the absence of ANP32B.
Table 4.
Loss of Anp32b reveals a moderate viability defect attributable to Anp32a
|
Anp32b+/+ |
Anp32b+/− |
Anp32b−/− |
|||||||
| Anp32a+/+ | Anp32a+/− | Anp32a−/− | Anp32a+/+ | Anp32a+/− | Anp32a−/− | Anp32a+/+ | Anp32a+/− | Anp32a−/− | |
| Mendelian | 10 | 20 | 10 | 20 | 41 | 20 | 10 | 20 | 10 |
| Expected | 15 | 30 | 15 | 23 | 45 | 23 | 3 | 6 | 3 |
| Observed* | 15 | 25 | 20 | 28 | 43 | 23 | 8 | 0 | 0 |
Numbers of double-mutant mice of the indicated genotypes arising from the intercrossing of Anp32a+/−;Anp32b+/− mice. “Mendelian” indicates the distribution for the indicated total number of mice under conditions of normal Mendelian segregation. “Expected” indicates the distribution for the Mendelian distributed numbers with integration of the observed rates of Anp32b lethality (Table 1). “Observed” indicates the actual numbers of mice of the noted genotypes obtained. Statistical probability is calculated against “Expected” ratios.
*P < 0.01 by χ2 analysis.
In contrast, loss of ANP32E did not further exacerbate the lethality associated with Anp32b deletion (Table 5). Although only one Anp32b−/−;Anp32e−/− mouse survived to weaning in this particular analysis, this rate was not statistically different from the survival rate expected for a mouse lacking only Anp32b−/− (Table 1). Furthermore, Anp32b−/−;Anp32e+/− mice exhibited a robust survival that stood in marked contrast to the fully penetrant lethality of Anp32b−/−;Anp32a+/− mice. When we attempted to generate triple mutants lacking ANP32A, ANP32B, and ANP32E, none were viable (as expected). Strikingly, a single functional Anp32b allele was sufficient to allow survival to weaning age in the mixed-bred background (Table S1), but a single functional Anp32a or Anp32e allele could not support mouse survival in the absence of ANP32B. These data indicate that there is a hierarchy of ANP32 protein functions in which ANP32B is the most important family member for embryogenesis, with ANP32A being of moderate importance and ANP32E being of least importance.
Table 5.
Loss of Anp32b does not reveal any defect attributable to Anp32e
|
Anp32b+/+ |
Anp32b+/− |
Anp32b−/− |
|||||||
| Anp32e+/+ | Anp32e+/− | Anp32e−/− | Anp32e+/+ | Anp32e+/− | Anp32e−/− | Anp32e+/+ | Anp32e+/− | Anp32e−/− | |
| Mendelian | 6 | 11 | 6 | 11 | 22 | 11 | 6 | 11 | 6 |
| Expected | 8 | 16 | 8 | 12 | 25 | 12 | 2 | 3 | 2 |
| Observed | 9 | 13 | 10 | 14 | 21 | 15 | 3 | 2 | 1 |
Number of double-mutant mice arising from the intercrossing of Anp32e+/−;Anp32b+/− mice. “Mendelian” indicates the distribution for the indicated total number of mice under conditions of normal Mendelian segregation. “Expected” indicates the distribution for the Mendelian distributed numbers with integration of the observed rates of Anp32b lethality (Table 1). “Observed” indicates the actual numbers of mice of the noted genotypes obtained. No statistically significant deviation from “Expected” values was observed using χ2 analysis.
Discussion
In this study, we provide evidence that ANP32B may be a useful prognostic indicator in human breast cancer and describe the generation and characterization of the ANP32B-deficient mouse.
We carried out a meta-analysis of ANP32B mRNA levels in human breast cancers using three independent studies in which the patients were subject to different exclusion criteria and treatment regimens (37–39). Although ANP32B was not highlighted in the original individual analyses, our examination of the combined data indicates that elevated ANP32B expression correlates with shortened patient survival. We hypothesize that ANP32B does not control tumorigenesis itself but rather increases, or is a marker of, the robustness of the resulting tumor cells; that is, ANP32B is elevated in tumor cells exhibiting increased proliferation and thus aggression. Consistent with this hypothesis, we found that Anp32b mRNA was up-regulated in murine cells exhibiting enhanced proliferation in tissue culture and in vivo.
Our report identifies an ANP32 mutation with a strong adverse effect on development. We have shown that null mutation of Anp32b in mice results in highly penetrant perinatal lethality in a mixed genetic background and in fully penetrant lethality in a pure C57BL/6 background. The specific cause of this lethality remains unclear because surviving mixed-bred Anp32b−/− mice suffered from a range of pathologies. We speculate that, during the course of evolution, the ANP32 proteins, and particularly ANP32B, took on functions that were less dispensable for embryogenesis. The nature of these functions remains obscure, as our data indicate that loss of ANP32B does not consistently affect one organ and that the majority of these animals die just after E17.5, when most murine organogenesis is complete.
Our histological examinations did not show any defects in vascular endothelium or smooth muscle, as might have been expected from previous reports on ANP32B functions (32, 40). However, our data regarding the incidence of hematomas in Anp32b−/− embryos do support a potential role for ANP32B in vascular development. Considering the recognized function of KLF5 in vasculogenesis (32), and the reported influence of ANP32B on KLF5-activated transcription, it is possible that the hematomas may have been due to defects in ANP32B-assisted, KLF5-activated transcription.
Our results confirm genetically that there is considerable functional overlap among ANP32 proteins. If different ANP32 proteins functioned in discrete cell types, or in different processes within the same cell, we would not have expected to see the synergistic effects that we observed in our compound mutants. However, our most dramatic finding is that mice carrying Anp32b mutations are sensitized to loss of Anp32a, a gene that has not previously been shown to have an effect on mouse development. These data establish that, contrary to previous assumptions, ANP32 family members are not all functionally redundant in vivo. Although mutations of multiple alleles of Anp32a or Anp32e are compatible with embryogenesis, at least one functional Anp32b allele must be present in any compound mutant for normal rates of survival. Thus, ANP32B is clearly the most important member of the ANP32 family, at least in mice.
The broad array of functions ascribed to various ANP32 family members in the literature suggests that certain reported biochemical functions may be artifactual. The data we have presented here, as well as in a previous publication (35), demonstrate that the Anp32 genes are not likely to be broadly involved in apoptosis. Furthermore, no ANP32-deficient cells have yet shown proliferation defects, implying that ANP32-mediated inhibition of cell cycle control phosphatases, such as PP2A, may be redundant or have a limited impact on cell proliferation. Instead, we speculate that ANP32 proteins play a more direct role in controlling gene expression, likely through their reported activities in chromatin regulation and/or selective mRNA transport. In any case, regardless of the ANP32 protein function under investigation, our results indicate that future research aimed at understanding the shared functions of ANP32 proteins should concentrate on ANP32B.
Materials and Methods
Prognostic Marker Identification.
Data were acquired from three publicly available datasets (37–39). The prognostic risk associated with particular gene expression was computed using application of the Cox proportional hazard regression model.
Northern Blotting.
The multiple murine tissue blot for Northern analysis was acquired from Zyagen (catalog no. MN-MT-2). Anp32b mRNA was detected by hybridizing to a labeled probe consisting of the EcoRI–BamHI fragment at the 5′ end of the ORF. Gapdh was detected by probing with a 214-bp fragment that hybridized to exons 5, 6, and 7.
Real-Time RT-PCR.
RNA was extracted from specified cell populations using a standard protocol and the RNeasy kit (Qiagen), quantified, and reverse-transcribed using a SuperScript II first-strand synthesis kit (Invitrogen). cDNA samples were then used as templates for quantitative real-time PCR (qPCR) using an ABI 7900HT detection system and SYBR Green (Applied Biosystems). Data were normalized to Actb (β-actin), Tbp mRNA, or Rn18S (18S rRNA). Primer sequences are provided in Table S2. Statistical significance of differences in normalized values were assessed by Student's t test.
Mice.
Mice were maintained under specified-pathogen free conditions in individually ventilated cages and fed a 5% irradiated meal. Anp32b−/− mice analyzed in this study were derived from two separate homologous recombinant clones and analyzed in approximately equal proportion for all experiments. Unless otherwise stated, analyses were performed in the mixed-bred 129ola;C57BL/6 genetic background. Statistical analyses for weights and longevity were performed using Student's t test and log-rank analysis, respectively.
Plasmids and Primers.
Sequences directing the expression of diphtheria toxin A in mouse ES cells were cloned into pBluescript (pgk-neo) to give the plasmid pBSneoDTA (35). Regions of the Anp32b gene adjacent to the targeted exons were cloned by high-fidelity PCR. Primers used to clone the upstream and downstream arms of homology into the XhoI site and XbaI site, respectively, are shown in Table S2.
Gene Targeting.
Targeting constructs were linearized and transfected by electroporation into E14K mouse ES cells as previously described (41, 42). Southern probes used to detect Anp32b genomic sequences were amplified from genomic ES cell DNA. PCR primer sequences for Anp32b flanking probe generation are presented in Table S2.
Genotyping.
For standard genotyping, PCR was performed on proteinase-treated biopsies. Primer sequences are provided in Table S2. Statistical analyses of progeny derived from intercrosses were performed using χ2 analysis.
Immunoblotting.
Mouse thymi were homogenized and lysed in buffer containing Nonidet P-40 detergent. Protein extracts (5 μg) were fractionated by SDS/PAGE, transferred to PVDF membranes, and probed with anti-ANP32B antibody (10843-1-AP; Proteintech) or anti–β-tubulin (loading control; Li-Cor Bioscience). Protein bands on blots were visualized using Li-Cor Odyssey standard protocols.
Cell Populations.
Primary MEFs were prepared from littermate Anp32b+/+, Anp32b +/−, and Anp32b −/− E14.5 embryos derived from timed interbreedings of Anp32b+/− mice. For transformed MEFs, primary MEFs (5 × 105 cells) were infected with a retrovirus generated from pLPC E1A/rasV12 using published techniques (43). Single-cell suspensions of total thymocytes were prepared from three sex-matched littermate pairs of Anp32b +/+ and Anp32b −/− mice (4–8 wk old). Single-cell suspensions derived from spleen or lymph nodes were treated with red blood cell lysis buffer (Sigma) to remove erythrocytes, and B and T cells were purified using the IMag cell separation system (Becton Dickinson). Briefly, total leukocytes were incubated with mouse anti-CD16/32 blocking antibodies, after which T cells were negatively selected using biotinylated anti-Ter119/B220/CD19/CD11b/Nk1.1/CD11c antibodies, and B cells were positively selected using biotinylated anti-CD19/B220 antibodies.
Apoptosis.
Apoptosis assays were performed as described previously (35, 44). Briefly, cells were exposed to apoptotic stimuli as detailed in Fig. S3 and cultured overnight in medium containing penicillin/streptomycin, 10% FBS, and l-glutamine. Viability was determined by propidium iodide exclusion and flow cytometry using a FACScaliber (BD Pharmingen).
Proliferation.
The growth of primary MEFs in culture was analyzed as previously described (35). Purified B cells were treated with 1 μg/mL LPS for 24 h. Purified T cells were stimulated by seeding on plates precoated with anti-CD3 and anti-CD28 antibodies as previously described (45). Statistical differences in cell growth were assessed using Student's t test.
Supplementary Material
Acknowledgments
We thank Arda Shahinian, Gordon Duncan, Jennifer Silvester, Jaselyn Becz-Ortiz, Low Mei Jun, and Melissa Madden for technical assistance and Mary Saunders for scientific editing of the manuscript. This research was partially supported by the Prostate Cancer Research Foundation of Canada, the Department of Defense (Grant PC020806), and the Singapore Millennium Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106211108/-/DCSupplemental.
References
- 1.Santa-Coloma TA. Anp32e (Cpd1) and related protein phosphatase 2 inhibitors. Cerebellum. 2003;2:310–320. doi: 10.1080/14734220310017212. [DOI] [PubMed] [Google Scholar]
- 2.Huyton T, Wolberger C. The crystal structure of the tumor suppressor protein pp32 (Anp32a): Structural insights into Anp32 family of proteins. Protein Sci. 2007;16:1308–1315. doi: 10.1110/ps.072803507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matilla A, Radrizzani M. The Anp32 family of proteins containing leucine-rich repeats. Cerebellum. 2005;4:7–18. doi: 10.1080/14734220410019020. [DOI] [PubMed] [Google Scholar]
- 4.Anisimov SV, et al. SAGE identification of gene transcripts with profiles unique to pluripotent mouse R1 embryonic stem cells. Genomics. 2002;79:169–176. doi: 10.1006/geno.2002.6687. [DOI] [PubMed] [Google Scholar]
- 5.Brody JR, et al. pp32 reduction induces differentiation of TSU-Pr1 cells. Am J Pathol. 2004;164:273–283. doi: 10.1016/S0002-9440(10)63117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kular RK, Cvetanovic M, Siferd S, Kini AR, Opal P. Neuronal differentiation is regulated by leucine-rich acidic nuclear protein (LANP), a member of the inhibitor of histone acetyltransferase complex. J Biol Chem. 2009;284:7783–7792. doi: 10.1074/jbc.M806150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puente LG, Carrière JF, Kelly JF, Megeney LA. Comparative analysis of phosphoprotein-enriched myocyte proteomes reveals widespread alterations during differentiation. FEBS Lett. 2004;574:138–144. doi: 10.1016/j.febslet.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Adegbola O, Pasternack GR. Phosphorylated retinoblastoma protein complexes with pp32 and inhibits pp32-mediated apoptosis. J Biol Chem. 2005;280:15497–15502. doi: 10.1074/jbc.M411382200. [DOI] [PubMed] [Google Scholar]
- 9.Hoffarth S, et al. pp32/PHAPI determines the apoptosis response of non-small-cell lung cancer. Cell Death Differ. 2008;15:161–170. doi: 10.1038/sj.cdd.4402256. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, et al. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science. 2003;299:223–226. doi: 10.1126/science.1076807. [DOI] [PubMed] [Google Scholar]
- 11.Mazroui R, et al. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J Cell Biol. 2008;180:113–127. doi: 10.1083/jcb.200709030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan W, et al. PHAPI/pp32 suppresses tumorigenesis by stimulating apoptosis. J Biol Chem. 2009;284:6946–6954. doi: 10.1074/jbc.M805801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schafer ZT, et al. Enhanced sensitivity to cytochrome c-induced apoptosis mediated by PHAPI in breast cancer cells. Cancer Res. 2006;66:2210–2218. doi: 10.1158/0008-5472.CAN-05-3923. [DOI] [PubMed] [Google Scholar]
- 14.Sun W, et al. Proliferation related acidic leucine-rich protein PAL31 functions as a caspase-3 inhibitor. Biochem Biophys Res Commun. 2006;342:817–823. doi: 10.1016/j.bbrc.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Amasaki H, et al. Distributional changes of BrdU, PCNA, E2F1 and PAL31 molecules in developing murine palatal rugae. Ann Anat. 2003;185:517–523. doi: 10.1016/s0940-9602(03)80116-4. [DOI] [PubMed] [Google Scholar]
- 16.Fukukawa C, Tanuma N, Okada T, Kikuchi K, Shima H. pp32/I-1(PP2A) negatively regulates the Raf-1/MEK/ERK pathway. Cancer Lett. 2005;226:155–160. doi: 10.1016/j.canlet.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 17.Sun W, et al. PAL31, a nuclear protein required for progression to the S phase. Biochem Biophys Res Commun. 2001;280:1048–1054. doi: 10.1006/bbrc.2000.4244. [DOI] [PubMed] [Google Scholar]
- 18.Costanzo RV, et al. Anp32e/Cpd1 regulates protein phosphatase 2A activity at synapses during synaptogenesis. Eur J Neurosci. 2006;23:309–324. doi: 10.1111/j.1460-9568.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Makkinje A, Damuni Z. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry. 1996;35:6998–7002. doi: 10.1021/bi960581y. [DOI] [PubMed] [Google Scholar]
- 20.Radrizzani M, et al. Differential expression of CPD1 during postnatal development in the mouse cerebellum. Brain Res. 2001;907:162–174. doi: 10.1016/s0006-8993(01)02351-4. [DOI] [PubMed] [Google Scholar]
- 21.Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ. Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 2004;23:2134–2145. doi: 10.1038/sj.emboj.7600210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fries B, et al. Analysis of nucleocytoplasmic trafficking of the HuR ligand APRIL and its influence on CD83 expression. J Biol Chem. 2007;282:4504–4515. doi: 10.1074/jbc.M608849200. [DOI] [PubMed] [Google Scholar]
- 23.Gallouzi IE, Brennan CM, Steitz JA. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. and partial retraction (2003) 9:1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itin C, Ulitzur N, Mühlbauer B, Pfeffer SR. Mapmodulin, cytoplasmic dynein, and microtubules enhance the transport of mannose 6-phosphate receptors from endosomes to the trans-golgi network. Mol Biol Cell. 1999;10:2191–2197. doi: 10.1091/mbc.10.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opal P, et al. Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesis. J Biol Chem. 2003;278:34691–34699. doi: 10.1074/jbc.M302785200. [DOI] [PubMed] [Google Scholar]
- 26.Kutney SN, Hong R, Macfarlan T, Chakravarti D. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression. J Biol Chem. 2004;279:30850–30855. doi: 10.1074/jbc.M404969200. [DOI] [PubMed] [Google Scholar]
- 27.Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- 28.Seo SB, et al. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem. 2002;277:14005–14010. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- 29.Tochio N, et al. Solution structure of histone chaperone ANP32B: Interaction with core histones H3-H4 through its acidic concave domain. J Mol Biol. 2010;401:97–114. doi: 10.1016/j.jmb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Cvetanovic M, et al. The role of LANP and ataxin 1 in E4F-mediated transcriptional repression. EMBO Rep. 2007;8:671–677. doi: 10.1038/sj.embor.7400983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loven MA, et al. A novel estrogen receptor alpha-associated protein alters receptor-deoxyribonucleic acid interactions and represses receptor-mediated transcription. Mol Endocrinol. 2004;18:2649–2659. doi: 10.1210/me.2003-0195. [DOI] [PubMed] [Google Scholar]
- 32.Munemasa Y, et al. Promoter region-specific histone incorporation by the novel histone chaperone ANP32B and DNA-binding factor KLF5. Mol Cell Biol. 2008;28:1171–1181. doi: 10.1128/MCB.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen SM, et al. Downregulation of ANP32B, a novel substrate of caspase-3, enhances caspase-3 activation and apoptosis induction in myeloid leukemic cells. Carcinogenesis. 2010;31:419–426. doi: 10.1093/carcin/bgp320. [DOI] [PubMed] [Google Scholar]
- 34.Opal P, et al. Generation and characterization of LANP/pp32 null mice. Mol Cell Biol. 2004;24:3140–3149. doi: 10.1128/MCB.24.8.3140-3149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reilly PT, et al. Generation and characterization of the Anp32e-deficient mouse. PLoS ONE. 2010;5:e13597. doi: 10.1371/journal.pone.0013597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buszczak M, et al. The carnegie protein trap library: A versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 38.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zippo A, De Robertis A, Bardelli M, Galvagni F, Oliviero S. Identification of Flk-1 target genes in vasculogenesis: Pim-1 is required for endothelial and mural cell differentiation in vitro. Blood. 2004;103:4536–4544. doi: 10.1182/blood-2003-11-3827. [DOI] [PubMed] [Google Scholar]
- 41.Okada H, et al. Generation and characterization of Smac/DIABLO-deficient mice. Mol Cell Biol. 2002;22:3509–3517. doi: 10.1128/MCB.22.10.3509-3517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruland J, et al. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc Natl Acad Sci USA. 2001;98:1859–1864. doi: 10.1073/pnas.98.4.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin AW, et al. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaugg K, et al. Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc Natl Acad Sci USA. 2007;104:3805–3810. doi: 10.1073/pnas.0611584104. and correction (2007) 104: 8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dufner A, et al. CARD6 is interferon inducible but not involved in nucleotide-binding oligomerization domain protein signaling leading to NF-kappaB activation. Mol Cell Biol. 2008;28:1541–1552. doi: 10.1128/MCB.01359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.