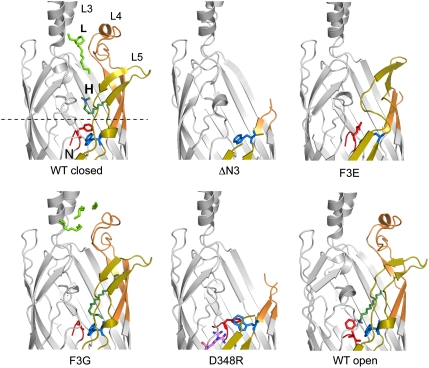
Fig. 3.
Effect of N-terminal mutations on the structure of EcFadL. Cartoons viewed from the side, shown in identical orientations, for closed wild-type EcFadL [Protein Data Bank (PDB) ID 1T16; ref. 6] and the mutants ΔN3, F3E, F3G, and D348R. In addition, the structure of a putative open wild-type channel is shown (PDB ID 1T1L; ref. 6). Loops L3–L5 are indicated in wild-type FadL. Detergent (green) bound in the low-affinity binding site is indicated with “L,” and that in the high-affinity binding site by “H.” Residue R348 in the D348R mutant is indicated with magenta sticks, with two conformations for the guanidinium group. The N terminus (residues G1–F3) is colored red, and W298 is colored blue. The dotted line in the wild-type protein corresponds to the center of the slabs through surfaces shown in Fig. 4.