Fig. 1.
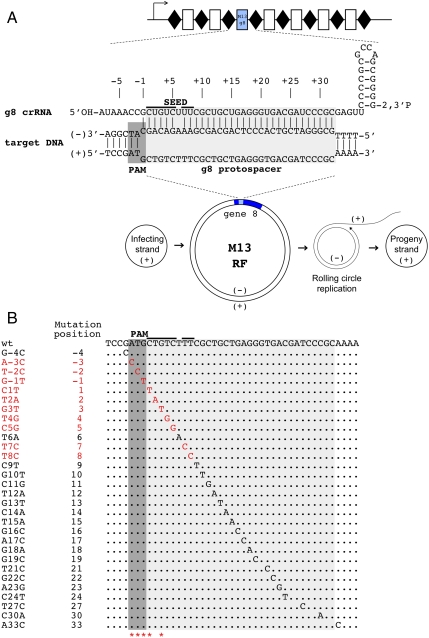
CRISPR/Cas mediated restriction of bacteriophage M13 requires intact PAM and PAM-proximal part of the protospacer. (A) An engineered plasmid-borne CRISPR cassette carrying a g8 spacer is schematically shown (Top). Rectangles indicate spacers, rhombi repeats. An arrow indicates the direction of transcription. Bottom) The life cycle of the M13 phage is schematically shown. Phage DNA enters the cell as a circular single-stranded DNA (“infecting” or “(+)” strand DNA). This strand is used as a template to create double-stranded “RF” of the phage genome (shown with gene 8, containing the g8 protospacer, highlighted with blue color). From the RF, the rolling circle replication of the phage genome is initiated, ultimately generating progeny (+) genome strands that are packaged in virions, The g8 protospacer and adjacent M13 sequence and crRNA containing the g8 spacer sequence are expanded above the RF intermediate. The structure shown (Top) forms when crRNA-guided Cascade complex recognizes double-stranded DNA containing the protospacer. (B) The sequence of the g8 protospacer, PAM, and the seed region is shown (Top). Below, point mutations selected as spontaneous escape phages on lawns of cells expressing g8 crRNA and also engineered by site-specific mutagenesis are shown. Mutations indicated with red color, led to escape, whereas mutations shown in black were restricted by CRISPR/Cas (the phage mutants had nonescape phenotype). Asterisks indicate positions of spontaneous escape mutations (see also Figs. S2A and S4). Additional point mutations obtained can be found in Fig. S2B.