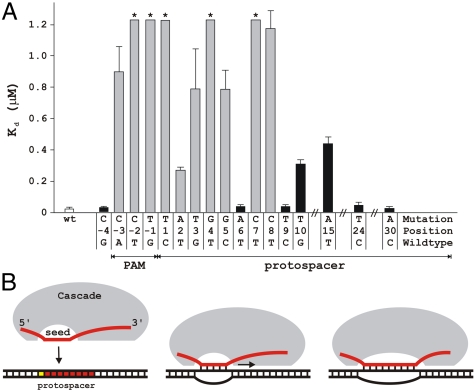
Fig. 3.
Mutations in the PAM sequence and mismatches in the seed region decrease target DNA binding affinity of the g8 crRNA-Cascade. (A) Dissociation constants (Kd) of escape mutant (gray bars) and nonescape mutant DNA (black bars) were determined by electrophoretic mobility shift assay. Asterisks indicate dissociation constants of these dsDNA targets that are greater than 1.2 μM. Error bars indicate standard deviations. (B) A model of R-loop formation between crRNA and double-stranded target DNA. Base pairing between the crRNA spacer and target DNA is initiated at crRNA seed sequence area and propagated in 5′ → 3′ direction over the complete protospacer region.