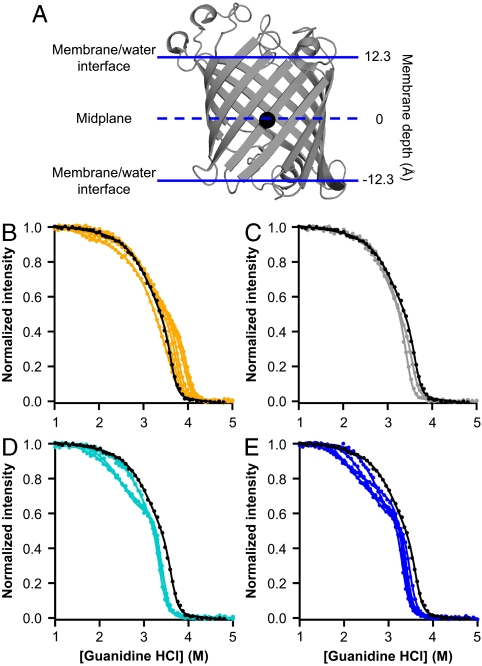
Fig. 1.
Outer membrane phospholipase A (OmpLA) as a host-guest system for measuring the membrane insertion energies of lipid-exposed amino acid residues. (A) Backbone of OmpLA rendered using PyMol (DeLano Scientific) and Protein Data Bank (PDB) ID code 1qd5 with the α-carbon of the alanine at sequence position 210 shown as a black sphere. Solid horizontal lines represent the boundaries of OmpLA’s transmembrane region. The dashed horizontal line represents the midplane of that region. (B–E) Example denaturation data for A210X sequence variants, each with a fit line from a three-state linear-extrapolation model. Data points represent tryptophan fluorescence emission measurements, normalized such that the emission from folded protein is 1 and the emission from unfolded protein is 0. Data for the wild-type OmpLA is shown in black in each panel. (B) Data for X = F, I, L, M, P, V, W, and Y are shown in orange. (C) Data for X = C and G are shown in gray. (D) Data for X = N, Q, S, and T are shown in teal. (E) Data for X = D, E, H, K, and R are shown in blue.