Abstract
Stem cell therapies offer the potential for repair and regeneration of cardiac tissue. To facilitate evaluation of stem cell activity in vivo, we created novel dual-reporter mouse embryonic stem (mES) cell lines that express the firefly luciferase (LUC) reporter gene under the control of the cardiac sodium–calcium exchanger-1 (Ncx-1) promoter in the background of the 7AC5-EYFP mES cell line that constitutively expresses the enhanced yellow fluorescent protein (EYFP). We compared the ability of recombinant clonal cell lines to express LUC before and after induction of cardiac differentiation in vitro. In particular, one of the clonal cell lines (Ncx-1-43LUC mES cells) showed markedly enhanced LUC expression (45-fold increase) upon induction of cardiac differentiation in vitro. Further, cardiac differentiation in these cells was perpetuated over a period of 2–4 weeks after transplantation in a neonatal mouse heart model, as monitored by noninvasive bioluminescence imaging (BLI) and confirmed via postmortem immunofluorescence and histological assessments. In contrast, transplantation of undifferentiated pluripotent Ncx-1-43LUC mES cells in neonatal hearts did not result in detectable levels of cardiac differentiation in these cells in vivo. These results suggest that prior induction of cardiac differentiation in vitro enhances development and maintenance of a cardiomyocyte-like phenotype for mES cells following transplantation into neonatal mouse hearts in vivo. We conclude that the Ncx-1-43LUC mES cell line is a novel tool for monitoring early cardiac differentiation in vivo using noninvasive BLI.
Introduction
Stem cell therapies hold great promise for the treatment of cardiovascular disease [1–3]. Basic and clinical research on cardiac applications for stem cells has intensified in recent years and much progress has been made [4–8], yet there is still a great deal of basic information about stem cells and their behavior that is lacking [9–11]. This is particularly true in vivo where it is difficult to assess cellular activity in the heart in real time. It would be helpful, therefore, to have better tools to track stem cell location and functional status over time in vivo.
Various in vivo molecular and cellular imaging techniques have been successfully applied for identifying and tracking transplanted stem cells in animal models [12–16]. Bioluminescence imaging (BLI), for example, uses light emission produced through the catalysis of luciferin by a luciferase (LUC) enzyme [16,17], and has proven useful for tracking and evaluating embryonic and adult stem cells in small animal models of heart disease [18–24]. Although BLI does not provide nearly the spatial resolution achieved with other imaging modalities such as magnetic resonance imaging, it is a promising technique that may be especially useful in small animal models because there is less tissue for the light to traverse than in larger animals and hence, less opportunity for signal loss or diminution due to scatter. By using different cellular promoters to drive LUC reporter gene expression, BLI can, in theory, provide information about the functional status and relative location of transplanted stem cells in vivo, but its use for evaluation of cellular differentiation status in vivo remains largely unexplored. Noninvasive evaluation of differentiation status could be helpful, for example, as an indicator of cardiac muscle regeneration growth in vivo.
In the present study, we used BLI to monitor differentiation and proliferation of mouse embryonic stem (mES) cells following transplantation into neonatal mouse hearts. To do this, we first created novel dual-reporter mES cell lines that express the firefly LUC reporter gene under the control of the cardiac Ncx-1 promoter. The LUC-expressing mES cells were derived from the recombinant EYFP mES cell line that constitutively expresses high levels of the enhanced yellow fluorescent protein (EYFP) [25]. Notably, EYFP expression is robust in the pluripotent mES cells and remains so as they differentiate into virtually any cell type, as previously demonstrated [25]. This is useful for identifying differentiating or differentiated mES cells in tissue sections from hearts that had received stem cell transplantations, but EYFP is not practical for in vivo cardiac imaging because the fluorescent signal is generally too weak for external detection. Thus, the utility of EYFP expression is presently limited to identification of the transplanted mES cells in postmortem tissue sections, while LUC expression permits evaluation of the transplanted mES cells over time in vivo.
Materials and Methods
Plasmids
The plasmids used in this study were kindly provided by the following researchers: Dr. Donald Menick, Medical University of South Carolina (Ncx-1-LUC) [26]; Dr. Brent French, University of Virginia (α-MHC-LUC) [27]; Dr. Shan Lu, University of Cincinnati (GAPDH-LUC) [28], and Dr. Karl Pfeifer, National Institute of Child Health and Development (MC1-Neo) [29]. Gene name abbreviations: Ncx-1, sodium–calcium exchanger-1; α-MHC, α-myosin heavy chain; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; and Neo, neomycin resistance gene.
Cell culture
Pluripotent 7AC5-EYFP R1-derived mES cells [25] were obtained from the American Tissue Culture Collection (ATCC SCRC-1033) and maintained as previously described [13]. Defined, iron-supplemented fetal bovine serum was obtained from Hyclone Labs (Logan, UT), and leukemia inhibitory factor (LIF) was procured from Chemicon, Inc. (Temecula, CA). Unless otherwise noted, all remaining media and supplements were obtained from Invitrogen (Carlsbad, CA).
Electroporation, colony selection, and clonal analysis
Electroporation and colony selection of mES cells were performed as previously described [29,30] with the following modifications. Approximately 50 μg of each plasmid DNA was linearized by restriction digestion, and re-purified to remove salts prior to electroporation. Four 60-mm plates of mES cells at optimal density (∼1 × 107 cells total) were washed once with PBS and treated with 0.05% Trypsin/EDTA. Cells were then dissociated into a single-cell suspension with gentle pipetting and the addition of 4 mL ES cell media to stop the reaction. The cells were pooled into a 15-mL conical tube and spun in the centrifuge to remove serum that could interfere with electroporation. The cell pellet was resuspended in 0.4 mL Hank's balanced salt solution (HBSS) containing linearized plasmids at 3:2 ratio for LUC:Neo vectors. The DNA-cell solution was then transferred to a sterile cuvette and electroporated using a Biorad Gene Pulser at 0.4 kV and 25 μFd. Electroporated cells were allowed to recover for 5 min and cells were transferred to fresh 60-mm plates containing mitomycin-inactivated MEFs. Cells were allowed to grow in normal culture media for 24 h and then selected with antibiotic G418 (Geneticin) at a concentration of 350 μg/mL. Colonies appeared at ∼7 days. The day prior to picking colonies, 24-well plates were pre-seeded with mitomycin-treated MEFs. Each colony was checked by microscopy for clear-defined ridges [30], and these were collected and trypsinized in a 24-well plate. The trypsinized cells were then transferred to fresh 24-well plates containing mitomycin-treated fibroblasts. After careful expansion for 2 passages, one batch was frozen for backup and the other batch was maintained for subsequent screening. The G418-resistant clonal cells were maintained on MEFs for 5 passages, and then grown without MEFs for subsequent testing.
LUC assays were performed using Glo-Lysis Buffer and the Steady-Glo LUC Assay System (Promega, Madison, WI). LUC activity was quantified directly on the multiwell plates using an In vivo Imaging System (IVIS-50; Caliper, Inc., Hopkinton, MA). Great care was taken to assay similar numbers of ES cells when comparing different clones and pluripotent versus cardiac-differentiated ES cells. Approximately 6–8 EBs (7 + 0 days) were seeded into each well of a 48-well plate, and ∼5 × 104 pluripotent cells were seeded into adjacent wells on the same plate on the following day (7 + 1 days). The cultures were incubated for an additional 48 h and the appearance of beating activity was confirmed in the cardiac-differentiated wells by visual inspection using phase-contrast microscopy prior to lysis and measurement of LUC activity at 7 + 3 days. Protein concentrations were determined using the DC Protein Assay (BioRad, Hercules, CA), and LUC activity was normalized to the total amount of protein per well.
To generate GAPDH-LUC EYFP mES cells, we screened 98 undifferentiated G418-resistant colonies for LUC expression in multiwell plates, and obtained 3 clones that showed stable expression. For α-MHC-LUC EYFP mES cells, we screened 182 G418-resistant colonies before and after induction of cardiac differentiation. Of these, 12 clones displayed LUC activity. Nine of these 12 were discontinued because of low or unstable expression following cardiac differentiation, while the other 3 clones were chosen for further characterization. The third cell line produced was stably transfected with the cardiac Ncx-1-LUC plasmid. Of 144 G418-resistant colonies, we initially found 23 that displayed LUC expression, and 9 of these continued to express LUC after secondary screening.
Induction of cardiac differentiation
Pluripotent (pluri) mES cells were induced to undergo cardiac differentiation (cardio) by the hanging-drop [31,32] (in vitro assays) or rotary shaker [33] (in vivo transplantation) methods using media conditions described previously [13]. In both cases, there was a 7-day suspension phase to develop embryoid bodies (EBs) followed by a 3-day adherent phase (7 + 3 days) when cardiac beating activity began to appear. All experiments using cardiac-differentiated cells in this study were performed with mES cells that had been induced to differentiate into beating cardiomyocytes for 7 + 3 days. Beating activity was confirmed by visual inspection using phase-contrast microscopy. Similar numbers of beating areas representing an estimated 2%–4% of the total cell population were observed, on average, for EBs generated by either the hanging-drop or rotary suspension methods. In addition, the size and morphology of the EBs appeared qualitatively similar in both groups.
Percoll density gradients
Cardiac-induced Ncx-1-43LUC mES cells were trypsinized at 7 + 3 days and centrifuged in a discontinuous gradient containing 0%, 25%, 50%, 100% Percoll™ at 400g for 35 min as previously described [34]. Identical gradients of this type containing density calibration beads were run in parallel for each of these experiments. Three fractions were collected as follows: (a) <1.06 g/mL, (b) 1.06–1.1 g/mL, and (c) ≥1.1 g/mL. Each of these fractions contained 2–3 mL. The fractionated cells were then resuspended, rinsed, and resuspended in ES culture media with serum. Cells from each fraction were seeded at 25,000 cells per well onto 4-well culture dishes. Bromodeoxyuridine (0.1 mM) was added to the culture media for the first 3 days to inhibit cell proliferation [35]. Beating activity was monitored by visual inspection using phase-contrast microscopy, and LUC activity was measured from freshly lysed cell extracts on the dish using BLI as described below.
Real-time quantitative polymerase chain reaction
The cDNA was prepared using Applied Biosystems™ High Capacity cDNA synthesis kit. We utilized 500 ng of total RNA for each cDNA synthesis reaction. The conditions for cDNA synthesis were followed per the protocol supplied with the cDNA synthesis kit. The cDNA was stored at −20°C until used for real-time PCR quantification. RT-qPCR conditions were standard for all samples. PCR primers were custom-designed and purchased from Operon (Huntsville, AL). All primers were diluted at a ratio of 1:10 or 1:20 based on optimal amplification of cDNA, determined by a relative standard curve for each primer set. In brief, the thermal cycling parameters were 95°C for 5 min, followed by 40–50 cycles of 95°C for 30 s (56°C or 58°C) for 30 s, 72°C for 45 s, and final extension at 72°C for 5 min, followed by a 4°C idle. RT-qPCR reactions were performed using IQ SYBR Green supermix obtained from BioRad, and run in the BioRad ICycler. The annealing temperatures for Ncx-1 and β-actin were 58°C and 56°C, respectively. At the end of each RT-qPCR experiment, the samples were analyzed by gel electrophoresis to confirm the presence of an appropriate size band (Ncx-1, 140 bp; β-actin, 420 bp). The primer sequences are as follows (5′–3′): mouse β-actin—forward: CATCACTATTGGCAACGAGC, reverse: ACGCAGCTCAGTAACAGTCC; mouse Ncx-1—forward: GACCAAGCAAGGAAGGCTGT, reverse: CGCAT AATGGTGAGGGCCACAG.
Cardiac injection
Pluripotent and cardiac-induced mES cells were trypsinized, centrifuged, and resuspended in HBSS at various concentrations. Neonatal (postnatal days 2–4) C57Bl/6 mice were anesthetized on ice for 2 min. The injection site was swabbed with 70% isopropyl alcohol before and after injection. The heart was visualized translucently in vivo using a fiber-optic light source projecting from the back of the neonatal thorax, and the cells were injected into the left ventricular muscle along the longitudinal axis with a 28G Tuberculin syringe (20 μL/injection). Peanut oil was then sparsely applied to the pup to ease maternal re-acceptance [36].
Bioluminescence imaging
Bioluminescence was measured using an In Vivo Imaging System 50 (IVIS™; Caliper, Hopkinton, MA) at 37°C. Mice were injected (150 mg/kg body weight, i.p.) with d-luciferin (Caliper) immediately prior to anesthesia. Mice were anesthetized with 2% isoflurane and imaged at 1- and 5-min exposure times. Bioluminescent signals were analyzed using Living Image® 2.50.2 software (Caliper) to identify regions of interest, quantitate light emission, and subtract background luminescence.
Histological and immunofluorescent staining
These procedures were performed as described previously [13,37].
Statistical analyses
Data are expressed as mean ± SEM unless indicated otherwise. Statistical comparisons (t-tests and one-way analysis of variance) and graphs were produced using Graphpad Prism™ 5 software (Graphpad Software, La Jolla, CA), with P < 0.05 required to reject the null hypothesis.
Results
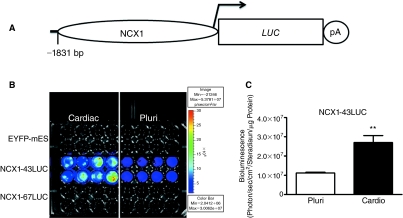
Our initial goal was to produce novel dual-reporter recombinant mES cell lines that express promoter-specific LUC as an in vivo reporter of cardiac differentiation in the background of a parental mES cell line that robustly expresses the EYFP reporter gene in all cell types [25]. To accomplish this, we used the cardiac Ncx-1 promoter (−1,831 bp) to drive LUC expression (Fig. 1A). For comparison, we also used GAPDH-LUC and α-MHC-LUC plasmids to generate recombinant mES cell lines. Characterization of several of the resulting clones is shown in Supplementary Fig. 1 (Supplementary materials are available online at www.liebertonline.com/scd), where it can be seen that most of the Ncx-1 promoter lines showed LUC activity that increased following induction of cardiac differentiation, while the other promoter constructs generated cells that showed either decreased or no change in LUC expression following induction of cardiac differentiation (Supplementary Fig. 1). Further screening of these LUC-expressing clones revealed the Ncx-1-43LUC clonal line to be an attractive candidate because it consistently produced stable baseline activity that increased significantly (∼240%, P < 0.01, n = 8) after induction of cardiac differentiation (Fig. 1B and 1C).
FIG. 1.
Comparison of luciferase (LUC) enzyme activity from the Ncx-1-43LUC clonal cell line in the pluripotent (Pluri) condition and following induction of cardiac differentiation (cardio) in vitro. (A) Schematic illustration of the Ncx-1-LUC construct used to transfect mouse embryonic stem (mES) cells. (B) Bioluminescence imaging (BLI) results from recombinant mES clones in the pluri or cardio states. Eight wells of each of the 3 cell lines indicated were evaluated in parallel for both conditions. (C) Quantitative assessment of the Ncx-1-43LUC results shown in panel B. **, P < 0.01. Color images available online at www.liebertonline.com/scd.
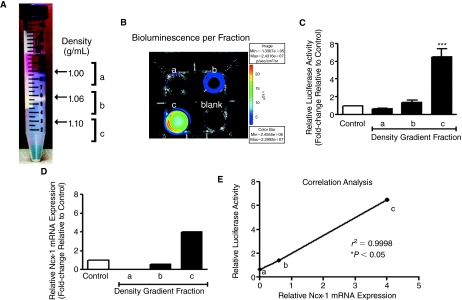
To enrich for cardiomyocytes, we fractionated the cell populations using Percoll density gradients [34] after induction of cardiac differentiation (7 + 3 days) (Fig. 2A). The vast majority (∼90%) of the cells were in fraction “b” (1.06–1.10 g/mL), which contained few or no cardiomyocytes. A thinner band of cell and cellular debris was found in fraction “a” (<1.06 g/mL). The remaining 3%–8% of the cell population was found in fraction “c” (>1.1 g/mL). Cells from each fraction were collected and cultured overnight. Contracting cardiomyocytes were predominantly found in fraction “c,” where the majority of the cells (>60%) displayed spontaneous contractile activities when assessed 24–48 h after plating. Relatively little beating activity was observed in fractions “a” and “b”. As with beating activity, LUC activity was also clearly strongest in fraction “c” (Fig. 2B and 2C). On a per-cell basis (25,000 cells/well), fraction “c” harbored roughly 10 times the amount of LUC activity compared with that found in fraction “a,” and nearly 5 times the amount in fraction “b,” which was similar to that observed for the unfractionated cardiac-induced mES cells. When compared with the amount of LUC activity in the unfractionated and undifferentiated (pluri) Ncx-1-43LUC mES cells (data not shown), the cardiomyocyte-enriched fraction “c” produced 45-fold more LUC activity, thereby further demonstrating that increased LUC activity is associated with cardiomyocyte differentiation in Ncx-1-43LUC mES cells.
FIG. 2.
Comparison of transgenic Ncx-1-LUC activity and endogenous Ncx-1 mRNA expression in cardiac-differentiated Ncx-1-43LUC cells. (A) Percoll density gradient fractionation of cardiac-differentiated Ncx-1-43 LUC mES cells. A large band of cells is visible just below the 1.06 g/mL density marker (fraction b) (note: densities were calculated using calibrated density marker beads in parallel gradients—not shown). This band contained ∼90% of the total number of cells loaded on the gradient. Smaller bands of cells were observed just below the 1.00 g/mL interface (fraction a) and, separately, at 1.1 g/mL (fraction c). Spontaneously contracting cardiomyocytes were predominantly found in fraction c, with few other cells. (B) Representative experiment showing LUC activity recorded using the IVIS from a 4-well dish with 25,000 cells per well from each designated fraction (a–c). One well did not contain cells (blank). (C) Relative LUC activity of cardiac-differentiated Ncx-1-43LUC mESCs before (control) and after fractionation. LUC activity is expressed relative to the control group. (D) Endogenous Ncx-1 mRNA was evaluated in fractions “a,” “b,” and “c” using RT-qPCR. Median values (n = 3) are shown normalized to actin and expressed relative to the unfractionated control cells. (E) Correlation analysis of relative Ncx-1-LUC activity and endogenous Ncx-1 mRNA expression in the different density gradient fractions. *P < 0.05; ***P < 0.001. Color images available online at www.liebertonline.com/scd.
To evaluate the specificity of the LUC activity produced from the Ncx-1-43LUC clonal cell line, we compared relative LUC activity and endogenous Ncx-1 mRNA expression levels from each fraction. RT-qPCR was performed to measure Ncx-1 mRNA levels in samples from each fraction relative to the unfractionated control samples. The overall pattern of Ncx-1 mRNA expression was similar to that produced from the LUC activity measurements in these samples (Fig. 2C and 2D), though the magnitude of the differences in expression between samples were less pronounced for the Ncx-1 mRNA changes compared with those observed for LUC activity. Nevertheless, there was a high degree of correlation (r2 = 0.9998) between relative LUC activity and Ncx-1 mRNA expression levels in the fractions analyzed (P < 0.05, n = 3) (Fig. 2E). These results indicate that the transgenic and endogenous Ncx-1 promoters behave similarly in that they both show marked increases in the cardiomyocyte-enriched fraction compared with the others, consistent with their expected patterns of expression [38,39].
To study the cardiac differentiation potential of the newly created dual-reporter mES cell lines in vivo, we transplanted pluripotent and cardiac-differentiated mES cells into neonatal mouse hearts via direct injection through the chest wall into left ventricular muscle. Measurement of LUC activity immediately after transplantation sometimes showed strong expression in the heart region, but often there was no expression observed or it disappeared in a matter of hours (Supplementary Fig. 2). Typically, however, stable in vivo bioluminescent signals appeared 4–8 days post-transplantation and could be observed over the remainder of the 24-day assessment period (Figs. 2–4). Although other clones showed differential LUC activity in pluripotent and cardiac-differentiated states, we found that the stability and robustness of the Ncx-1-43LUC mES cell line was most useful for in vivo bioluminescent imaging (BLI) compared with other clones analyzed (data not shown). Thus, we have used Ncx-1-43LUC mES cells for all subsequent experiments described in this report.
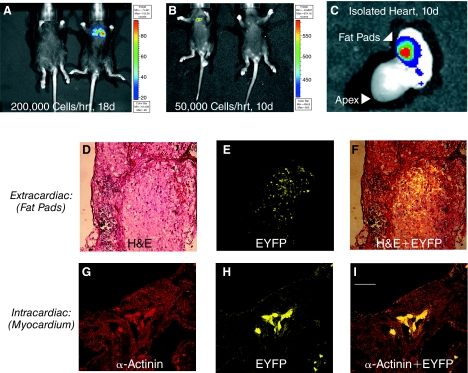
FIG. 4.
Evaluation of pluripotent Ncx-1-43LUC mES cell fate following transplantation into the neonatal mouse heart model. (A) Example of mES cells that spread to the lungs, as imaged 18 days after transplantation of 200,000 cells. (B) Example of mES cells that remained clustered in the cardiac region 10 days after transplantation of 50,000 cells. (C) Ex vivo imaging of the heart from the mouse shown in panel B. In this example, the signal appeared to emanate from the fat pad near the base of the heart, and this was confirmed by histological evaluation such as that shown in D–F. (D) Hematoxylin and eosin (H&E) staining of extracardiac tissue harboring mES cells. (E) EYFP expression in a serial section of extracardiac tissue adjacent to that shown in panel A. Scale bar, 100 μm. (F) Overlay of EYFP and H&E staining shown in panels D and E. (G–I) Intracardiac identification of transplanted mES cells in myocardial tissue. (G) Tissue sections from hearts that had received stem cell transplants were immunofluorescently labeled with an antibody that recognizes sarcomeric α-actinin and visualized using a Texas Red-conjugated secondary antibody. (H) Enhanced yellow fluorescent protein (EYFP) expression. (I) Overlay of α-actinin and EYFP staining in the same tissue section. Scale bar, 25 μm. Color images available online at www.liebertonline.com/scd.
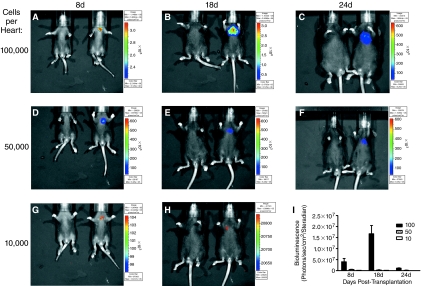
To determine an optimal number of cells to use per injection for BLI, we assessed in vivo bioluminescence activity following injection of various numbers of pluripotent Ncx-1-43LUC-expressing mES cells (Fig. 3). Injection of 100,000 or more cells led to robust bioluminescence expression that displayed peak activity at the 18-day time point, but then decreased precipitously over the next 6 days. Injection of 50,000 cells per heart resulted in more consistent bioluminescent signals that also appeared to remain more confined to the cardiac region rather than spreading over the chest area as observed with 100,000 cells (compare panels B and E, and C and F, Fig. 3). The minimum number of cells injected that were needed to observe a bioluminescent signal in vivo was 10,000 per heart, which produced a weak but measurable signal at 8 and 18 days post-transplantation (Fig. 3G and 3H). By 24 days post-transplantation, bioluminescent signals were no longer detected in the mice receiving only 10,000 cells per heart (data not shown).
FIG. 3.
Time course assessment of bioluminescence in vivo following transplantation of varying amounts of pluripotent Ncx-1-43LUC mES cells into neonatal mouse hearts. (A–C) BLI results following transplantation of 100,000 cells per heart at 8, 18, and 24 days post-transplantation. (D–F) BLI results following transplantation of 50,000 cells per heart at 8, 18, and 24 days post-transplantation. (G and H) BLI results following transplantation of 10,000 cells per heart at 8 and 18 days post-transplantation. In each panel, the mouse on the right had received the indicated cell transplantation while the mouse on the left served as a vehicle-injected control. The substrate, d-luciferin was administered to control and cell-treated mice ∼5 min prior to BLI. (I) Quantitative assessment of the results shown in A–H (n = 4–8/group). Key: 100 = 100,000 cells/injection (filled bars), 50 = 50,000 cells/injection (hatched bars), and 10 = 10,000 cells/injection (open bars). Color images available online at www.liebertonline.com/scd.
Bioluminescence remained clearly evident, however, in mice receiving 50,000 or more cells per heart through the entire 24-day assessment period, though the signal often decreased in intensity and became more diffuse at 24 days compared with 18 days post-transplantation. This trend was also observed using pluripotent GAPDH-6LUC and α-MHC-4LUC cells (Supplementary Fig. 3). Notably, tumor-like formations were observed in >80% of mice that were injected with 100,000 or more pluripotent mES cells per heart, irrespective of which promoter-LUC clone was used. Examples of this are shown in Figure 4. In one case, injection of 200,000 cells led to an apparent spread to lungs (Fig. 4A), as confirmed by postmortem tissue histological examinations. In another case when only 50,000 cells were injected, expression appeared confined to the heart region, but was later found to be located in the fat pads on the outside of the heart (Fig. 4B and 4C). Histological examinations showed that extracardiac concentrations of transplanted mES cells were commonly found near the heart (Fig. 4D–4F). In some mice, the transplanted pluripotent mES cells were found within both atrial and ventricular myocardium. They did not, however, appear to develop myocardial characteristics because they did not significantly express the myocyte marker protein, sarcomeric α-actinin. Thus, little or no sarcomeric structure was observed in the EYFP+ cells following transplantation of undifferentiated mES cells (Fig. 4G–4I), thereby indicating that they did not develop cardiomyocyte characteristics (morphology and structure) under these conditions.
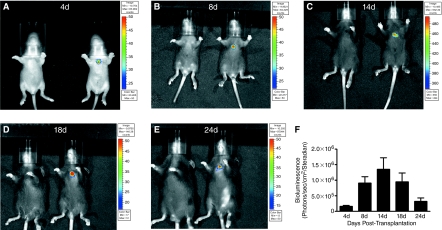
To improve the likelihood of cardiac differentiation and development of transplanted mES cells in vivo, we initiated cardiac differentiation of these cells in vitro en masse using the rotary suspension method [33]. As shown in Figure 5, transplantation of 50,000 cardiac-induced Ncx-1-43LUC mES cells resulted in relatively stable expression in the cardiac region in vivo. Expression was detected as early as 4 days post-transplantation (earliest time point measured) and remained clearly evident in the heart region through 24 days post-transplantation. Expression was highest between 8 and 18 days, with peak bioluminescence observed at 14 days post-transplantation. Tumor-like formations were still found in this group, but with reduced frequency (<25%) compared with the injections using 100,000 or more pluri cells (>80%).
FIG. 5.
Time course of in vivo bioluminescence activity in the developing mouse heart following transplantation of 50,000 Ncx-1-43LUC mES cells that had been induced to differentiate into beating cardiomyocytes (7 + 3 days) in vitro. (A–E) In vivo BLI at 4, 8, 18, and 24 days post-transplantation. Representative mice are shown for each time point, with the mouse on the left serving as the negative control (vehicle injection only) for the mouse on the right, which had been injected with the mES cells. (F) Graph of BLI data collected over the indicated time course (n = 7–16/time point). Color images available online at www.liebertonline.com/scd.
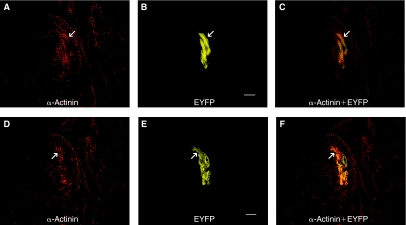
Postmortem immunofluorescent histological assessments confirmed that the cardiac-differentiated mES cells were found within the myocardium. Co-staining for the myocyte marker, sarcomeric α-actinin, and the transplanted cell marker, EYFP, was clearly observed, as shown in Figure 6. In this example, confocal fluorescent micrographs from 2 different optical planes of the same tissue section are shown in the top (panels A–C) and bottom (panels D–F) of Figure 6. In both cases, some of the EYFP cells display the ladder-like sarcomeric staining pattern that is characteristic of ventricular cardiac muscle cells. Further, the transplanted stem cells appear to be aligned in parallel and are morphologically similar to the endogenous ventricular myocytes found in adjacency. Several additional examples of this type of co-staining are provided in Supplementary Figure 4. These results demonstrate that transplanted cardio mES cells can develop and/or maintain a cardiomyocyte-like phenotype in the heart over a period of several weeks in vivo.
FIG. 6.
Dual fluorescent staining of transplanted Ncx-1-43LUC mES cells that had been differentiated into beating cardiomyocytes (7 + 3 days) in vitro. Each row of panels represents images from a single optical plane through ventricular myocardium stained with the muscle-specific marker, sarcomeric α-actinin (A and D), EYFP (B and E), or an overlay of α-actinin and EYFP (C and F). The staining was performed 14 days post-transplantation. Arrows indicate examples of EYFP+ mES cells that had developed sarcomeric structures as evidenced by the ladder-like staining pattern achieved from α-actinin staining in these sections. Scale bars, 10 μm. Color images available online at www.liebertonline.com/scd.
Based on the number of EYFP-labeled (EYFP+) cells in the tissue sections taken from these hearts, we estimated that the maximum number of mES-derived cells found in the heart 2 weeks after transplantation was ∼1.5 × 104 cells, and that fewer than half of them co-expressed the cardiomyocyte marker, sarcomeric α-actinin. Still, this was an “improvement” compared with the pluri transplantations where we did not find any cardiomyocyte-like EYFP+ cells.
To help relate BLI data with the number of cells present in vivo, we generated standard curves in vitro using defined numbers of undifferentiated Ncx-1-43LUC cells (see Supplementary Fig. 5). We used these curves as guidelines to estimate the number of “pluri-equivalent” cells present in vivo given a certain BLI reading. For example, a BLI reading of 5 × 106 photons/s/cm2/steradian predicts that there will be at least 1 × 105 “pluri-equivalent” cells present. The in vitro BLI signals do not have to traverse any mouse tissues, so it is anticipated that higher numbers of cells were needed to generate similar signal intensities in vivo because they would be diminished by tissue absorption and scatter, especially as the animals get older and bigger. This caveat notwithstanding, the standard curves are meant to serve simply as guidelines for estimating the minimum number of cells likely to be present in order to generate a BLI signal of a certain magnitude in vivo. Of note, the number of EYFP+ cells per mouse was highest when tumor-like formations were observed. Because of the high density of EYFP+ cells in these cases, it was difficult to obtain an accurate count of the EYFP+ cells per mouse, but we estimate with confidence that it was well in excess of 1 × 105 EYFP+ cells.
In contrast, peak BLI readings for the cardio mESCs were ∼1.3 × 106 photons/s/cm2/steradian at 14 days, which would predict a minimum of ∼3 × 104 “pluri-equivalent” Ncx-1-43LUC cells per heart. Actual EYFP+ cell counts from histological sections taken from hearts and surrounding tissues, however, indicated that a maximum of ∼1.5 × 104 EYFP+ cells were found at 14 days post-transplantation. These results suggest that the cardiac-differentiated mESCs continued to glow “brighter” than the undifferentiated “pluri” mESCs when similarly transplanted; otherwise, there should have been at least 2–3 times that many cells present in order to generate an equivalent BLI signal in vivo. Thus, these findings strongly support the premise that Ncx-1-43LUC cells can serve as an indicator of cardiac differentiation status using noninvasive BLI in vivo.
Discussion
In this study, we have described the production of novel dual-reporter mES cell lines and their application for in vivo BLI in the developing mouse heart. Using the EYFP mES cells created by Hadjantonakis et al. [25] as our starting material, we generated and characterized new clonal cell lines that stably express the LUC reporter under the control of different cellular promoters. In particular, the Ncx-1-43LUC EYFP mES cells represent a promising cell line for evaluation of cardiac differentiation status in vivo because it produces measurable baseline LUC activity that is strongly induced following differentiation into beating cardiomyocytes.
Most of the Ncx-1-LUC clonal lines displayed increased amounts of LUC activity when differentiated into beating cardiomyocytes when compared with that observed in the undifferentiated state. The best characterized of these clones is the Ncx-1-43LUC EYFP mES cell line, which was used for most of the in vivo BLI assessments in this study because of its stable basal level of LUC expression that becomes significantly enhanced following cardiac differentiation. In particular, the Ncx-1-43LUC EYFP mES cells represent a promising cell line for evaluation of cardiac differentiation status in vivo because it produces measurable baseline LUC activity that is strongly induced following differentiation into beating cardiomyocytes. The fact that endogenous Ncx-1 gene expression followed a similar pattern helps to support the idea that the transgenic Ncx-1-driven LUC expression faithfully reported endogenous expression patterns. Both clearly became elevated following differentiation of pluripotent Ncx-1-43LUC mESCs into cardiomyocytes.
Density gradient fractionation is an established method for cardiomyocyte enrichment using these types of systems [40,41], and our results demonstrate that enhanced expression levels of both endogenous and transgenic Ncx-1 were associated with the cardiomyocyte-enriched fraction. While this does not prove that Ncx-1 expression is selectively up-regulated in cardiomyocytes, these observations are consistent with earlier studies showing that low levels of Ncx-1 are expressed in pluripotent mES cells [42] and during gastrulation in vivo [38]. Importantly, they are also consistent with previous work showing that Ncx-1 expression increases dramatically in cardiomyocytes during early stages of cardiac development in vivo [38,39].
The relatively low percentage of stable clones obtained in our screening process likely derives from where and how the electroporated linearized plasmid DNA integrated into the mouse genome for each clonal line and the fact that the neomycyin resistance gene and the LUC reporter gene were located on separate plasmids. Since these are non-homologous random integration events, the resulting LUC activity will vary greatly depending upon nearby genomic DNA sequence, as has been commonly observed in other studies [43–45]. We did not directly examine the site of integration in our clones, but we did perform functional screening of the clones. So far, the most useful clone to emerge is the Ncx-1-43LUC cell line because of its intrinsic brightness and ability to display enhanced LUC activity upon cardiac differentiation.
In contrast, clones generated using either the GAPDH-LUC or α-MHC-LUC plasmids did not display increased LUC activity following cardiac differentiation. Those generated with GAPDH-LUC generally showed similar or slightly decreased amounts of LUC expression when differentiated into beating cardiomyocytes compared with undifferentiated controls (Supplementary Fig. 1). This was expected from a “housekeeping” promoter like GAPDH. The decreased expression associated with some of the α-MHC-LUC clones in the cardiac-differentiated state was somewhat of a surprise since this promoter is typically active in cardiomyocytes, but the relatively short (353 bp) promoter used for this construct was apparently insufficient for this purpose despite the fact that it contained key genetic elements important for myocyte expression [46]. Of the constructs and resulting clonal mES cell lines tested thus far, Ncx-1-43 was clearly the most promising as an evaluative tool to examine cardiac differentiation in vivo because of its strong induction in cardiomyocytes derived from differentiating mES cells in vitro.
The first assessment parameter that we evaluated in vivo was to determine the optimal number of recombinant LUC-expressing mES cells to transplant into the neonatal mouse heart model, defined as the minimum number of cells required to produce a BLI signal in vivo with relatively low risk of tumor-like growths of mES cells developing. Our results suggest that transplantation of 100,000 or more cells has a high probability of tumor-like formations, and this was true for all cell lines tested. Further, both pluri and cardio mES cells produced tumor-like formations when 100,000 or more of them were transplanted into the neonatal mouse heart. Consequently, we can reasonably conclude that using 100,000 or more mES cells leads to undesired side effects in this model, similar to that seen in other studies where pluripotent mES cells have been transplanted into adult mouse hearts in vivo [40,41].
On the other hand, if too few cells are transplanted into the heart, then it becomes difficult to visualize them in vivo using BLI, though there appears to be much lower risk of tumor-like formations appearing when using fewer cells [41]. We found injection of 10,000 cells per heart was the lower limit for subsequent detection of these cells in vivo using BLI techniques. This limit is dependent on the particular cell line used. For example, the GAPDH-6LUC and α-MHC-4LUC cells required 50,000–100,000 cells per heart in order to achieve a measurable BLI signal. This is likely due to a number of factors, including promoter strength, chromosomal context of the recombination site, and differentiation status of the cells. For most of our experiments with Ncx-1-43LUC EYFP mES cells, we used 50,000 cells per heart because these generated stronger and more consistent in vivo BLI signals (compared with 10,000 cells/heart) without necessarily producing tumor-like growths. Thus, another conclusion that can be drawn from these data is that injection of 10,000–50,000 Ncx-1-43LUC EYFP mES cells per heart was an effective number of cells for in vivo BLI assessments in this neonatal mouse model.
In the pluri state, we found little evidence for subsequent cardiomyocyte differentiation in transplanted mES cells, regardless of which cell line was used or how many cells were transplanted per heart. We could readily identify the transplanted mES cells in postmortem heart sections by strong EYFP expression, but found no clear examples of co-expression of EYFP with myocardial markers following pluri mES cell transplantations. While we cannot rule out the possibility that a few of the transplanted pluri mES cells may have differentiated into cardiomyocytes in vivo, it appears that this is a relatively rare event that is consistent with what other investigators have observed using similar ES cell transplantation strategies [40,41].
One key finding from the present study is that prior induction of cardiac differentiation appeared to promote the development and/or maintenance of a cardiomyocyte-like phenotype of the transplanted mES cells over a period of 2–4 weeks in vivo. This finding is consistent with that reported by Kolossov et al. [47], though the present study is the first to utilize imaging techniques to identify and quantify mES cellular differentiation activity over time in vivo. Further, mES cells identified in ventricular myocardium appeared to have aligned themselves in parallel with existing myocardium. While we do not yet know if they had become electromechanically coupled to existing myocytes, they did appear to be positioned in a manner consistent with physical integration with endogenous myocardium. Additional testing is required to determine if these cells are functionally coupled with surrounding myocardial cells in the host heart. At this point, however, we can conclude that in vitro differentiation of mES cells into beating cardiomyocytes prior to transplantation led to an improved ability of the transplanted mES cells to acquire and/or maintain cardiomyocyte-like characteristics in vivo.
One of the confounding issues that we and others face with these types of transplantation experiments is the significant loss of cells during or shortly after the transplantation procedure. In most cases, we found relatively few mES cells in host hearts compared with the relatively large numbers of cells that were delivered during the transplantation. While immune responses may account for some of this loss over time [41,48], there appears to be a more rapid loss that probably occurs during the transplantation procedure itself or shortly afterward, possibly due to mechanical forces [40,49,50]. This is partly because the neonatal myocardium has relatively thin walls that are rapidly beating as we were injecting the cells through the chest wall. Inevitably, many of the cells likely penetrated the heart wall and were lost in circulation or were deposited in nearby tissues such as cardiac fat pads, thymus, or lung. This appeared to be true regardless of whether pluri or cardio mES cells were used, and thus likely reflects a need for improved delivery strategies and techniques rather than issues concerning the differentiation status of the transplanted cells.
Although we plan to ultimately test these novel promoter-specific LUC-expressing mES cell lines in more clinically relevant adult cardiac injury models, the neonatal mouse heart model described here has proven useful as an initial in vivo screening tool for evaluation of novel recombinant mES reporter cell lines following their transplantation into the heart. This model offers several advantages over the adult model: (i) direct myocardial injection is feasible through the chest wall without the need to perform technically challenging open chest survival surgeries, (ii) the relatively small size of the neonate facilitates in vivo BLI because there is less tissue for the light emission to traverse than in adults, (iii) unlike adults, neonatal myocardium is still growing and developing and so, in theory, should provide a more conducive environment for differentiation and development of transplanted stem cells to become myocardial cells, (iv) the lack of an induced injury minimizes complications from remodeling and scar tissue formations, and (v) because of all of the above listed reasons, neonatal experiments can generally be performed more quickly and less expensively than analogous types of experiments using adult cardiac injury models. Thus, the neonatal model described here can serve as an effective in vivo screening tool to determine which reporter mES cell lines and in vitro differentiation/selection strategies have the ability to most effectively differentiate and develop into functional myocardial tissue in vivo. Future studies will need to evaluate these cells in an adult mouse model of myocardial infarction.
Summary
In summary, we have described the generation of recombinant dual-reporter mES cell lines that have the ability to provide useful information about the location, proliferation, and differentiation of transplanted stem cells in the heart. Using noninvasive BLI techniques, we were able to measure LUC expression in vivo, and identify the transplanted mES-derived cells via EYFP expression in situ. These results indicate that prior induction of cardiac differentiation in vitro enhances development and/or maintenance of a cardiomyocyte-like phenotype for mES cells following transplantation into neonatal mouse hearts in vivo. Further, we have shown that the neonatal mouse heart model provides an efficient and informative system to evaluate the behavior of these novel stem cell lines in vivo. This study represents a novel use of BLI to directly evaluate the differentiation status of transplanted stem cells in the developing heart. Future studies will seek to selectively isolate cardiomyocyte-enriched populations of these cells and further characterize their potential to repair/regenerate myocardium using a variety of in vitro and in vivo model systems.
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH to S.N.E (R01-HL078716) and a postdoctoral fellowship from the American Heart Association to D.G.T. (0625631B). The authors thank Ms. Tu-Suong Nguyen for technical assistance with the study.
Author Disclosure Statement
No competing financial interests exist.
References
- 2.Barile L. Messina E. Giacomello A. Marbán E. Endogenous cardiac stem cells. Prog Cardiovasc Dis. 2007;50:31–48. doi: 10.1016/j.pcad.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 1.Anversa P. Leri A. Rota M. Hosoda T. Bearzi C. Urbanek K. Kajstura J. Bolli R. Concise review: stem cells, myocardial regeneration, and methodological artifacts. Stem Cells. 2007;25:589–601. doi: 10.1634/stemcells.2006-0623. [DOI] [PubMed] [Google Scholar]
- 3.Rubart M. Field LJ. Cardiac regeneration: repopulating the heart. Annu Rev Physiol. 2006;68:29–49. doi: 10.1146/annurev.physiol.68.040104.124530. [DOI] [PubMed] [Google Scholar]
- 4.Bearzi C. Rota M. Hosoda T. Tillmanns J. Nascimbene A. De Angelis A. Yasuzawa-Amano S. Trofimova I. Siggins RW. Lecapitaine N. Cascapera S. Beltrami AP. D'Alessandro DA. Zias E. Quaini F. Urbanek K. Michler RE. Bolli R. Kajstura J. Leri A. Anversa P. Human cardiac stem cells. Proc Natl Acad Sci USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kattman SJ. Adler ED. Keller GM. Specification of multipotential cardiovascular progenitor cells during embryonic stem cell differentiation and embryonic development. Trends Cardiovasc Med. 2007;17:240–246. doi: 10.1016/j.tcm.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Penn MS. Mangi AA. Genetic enhancement of stem cell engraftment, survival, and efficacy. Circ Res. 2008;102:1471–1482. doi: 10.1161/CIRCRESAHA.108.175174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segers VF. Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 8.Yang L. Soonpaa MH. Adler ED. Roepke TK. Kattman SJ. Kennedy M. Henckaerts E. Bonham K. Abbott GW. Linden RM. Field LJ. Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 9.Braun T. Martire A. Cardiac stem cells: paradigm shift or broken promise? A view from developmental biology. Trends Biotechnol. 2007;25:441–447. doi: 10.1016/j.tibtech.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Murry CE. Reinecke H. Pabon LM. Regeneration gaps: observations on stem cells and cardiac repair. J Am Coll Cardiol. 2006;47:1777–1785. doi: 10.1016/j.jacc.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Passier R. van Laake LW. Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 12.Bara C. Ghodsizad A. Niehaus M. Makoui M. Piechaczek C. Martin U. Warnecke G. Karck M. Gams E. Klein HM. Haverich A. Ruhparwar A. In vivo echocardiographic imaging of transplanted human adult stem cells in the myocardium labeled with clinically applicable CliniMACS nanoparticles. J Am Soc Echocardiogr. 2006;19:563–568. doi: 10.1016/j.echo.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Ebert SN. Taylor DG. Nguyen HL. Kodack DP. Beyers RJ. Xu Y. Yang Z. French BA. Noninvasive tracking of cardiac embryonic stem cells in vivo using magnetic resonance imaging techniques. Stem Cells. 2007;25:2936–2944. doi: 10.1634/stemcells.2007-0216. [DOI] [PubMed] [Google Scholar]
- 14.Hinds KA. Hill JM. Shapiro EM. Laukkanen MO. Silva AC. Combs CA. Varney TR. Balaban RS. Koretsky AP. Dunbar CE. Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allows magnetic resonance imaging of single cells. Blood. 2003;102:867–872. doi: 10.1182/blood-2002-12-3669. [DOI] [PubMed] [Google Scholar]
- 15.Rogers WJ. Meyer CH. Kramer CM. Technology insight: in vivo cell tracking by use of MRI. Nat Clin Pract Cardiovasc Med. 2006;3:554–562. doi: 10.1038/ncpcardio0659. [DOI] [PubMed] [Google Scholar]
- 16.Wu JC. Chen IY. Sundaresan G. Min JJ. De A. Qiao JH. Fishbein MC. Gambhir SS. Molecular imaging of cardiac cell transplantation in living animals using optical bioluminescence and positron emission tomography. Circulation. 2003;108:1302–1305. doi: 10.1161/01.CIR.0000091252.20010.6E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu JC. Inubushi M. Sundaresan G. Schelbert HR. Gambhir SS. Optical imaging of cardiac reporter gene expression in living rats. Circulation. 2002;105:1631–1634. doi: 10.1161/01.cir.0000014984.95520.ad. [DOI] [PubMed] [Google Scholar]
- 18.Cao F. Lin S. Xie X. Ray P. Patel M. Zhang X. Drukker M. Dylla SJ. Connolly AJ. Chen X. Weissman IL. Gambhir SS. Wu JC. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao F. Wagner RA. Wilson KD. Xie X. Fu JD. Drukker M. Lee A. Li RA. Gambhir SS. Weissman IL. Robbins RC. Wu JC. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE. 2008;3:e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakamata Y. Murakami T. Kobayashi E. “Firefly rats” as an organ/cellular source for long-term in vivo bioluminescent imaging. Transplantation. 2006;81:1179–1184. doi: 10.1097/01.tp.0000203137.06587.4a. [DOI] [PubMed] [Google Scholar]
- 21.Huang M. Chan DA. Jia F. Xie X. Li Z. Hoyt G. Robbins RC. Chen X. Giaccia AJ. Wu JC. Short hairpin RNA interference therapy for ischemic heart disease. Circulation. 2008;118(14 Suppl):S226–S233. doi: 10.1161/CIRCULATIONAHA.107.760785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z. Wu JC. Sheikh AY. Kraft D. Cao F. Xie X. Patel M. Gambhir SS. Robbins RC. Cooke JP. Wu JC. Differentiation, survival, and function of embryonic stem cell derived endothelial cells for ischemic heart disease. Circulation. 2007;116(11 Suppl):I46–I54. doi: 10.1161/CIRCULATIONAHA.106.680561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheikh AY. Lin SA. Cao F. Cao Y. van der Bogt KE. Chu P. Chang CP. Contag CH. Robbins RC. Wu JC. Molecular imaging of bone marrow mononuclear cell homing and engraftment in ischemic myocardium. Stem Cells. 2007;25:2677–2684. doi: 10.1634/stemcells.2007-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Bogt KE. Sheikh AY. Schrepfer S. Hoyt G. Cao F. Ransohoff KJ. Swijnenburg RJ. Pearl J. Lee A. Fischbein M. Contag CH. Robbins RC. Wu JC. Comparison of different adult stem cell types for treatment of myocardial ischemia. Circulation. 2008;118(14 Suppl):S121–S129. doi: 10.1161/CIRCULATIONAHA.107.759480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadjantonakis AK. Macmaster S. Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes KV. Cheng G. Dawson MM. Menick DR. Cloning of cardiac, kidney, and brain promoters of the feline ncx1 gene. J Biol Chem. 1997;272:11510–11517. [PubMed] [Google Scholar]
- 27.Prasad KM. Xu Y. Yang Z. Toufektsian MC. Berr SS. French BA. Topoisomerase inhibition accelerates gene expression after adeno-associated virus-mediated gene transfer to the mammalian heart. Mol Ther. 2007;15:764–771. doi: 10.1038/sj.mt.6300071. [DOI] [PubMed] [Google Scholar]
- 28.Lu S. Gu X. Hoestje S. Epner DE. Identification of an additional hypoxia responsive element in the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Biochim Biophys Acta. 2002;1574:152–156. doi: 10.1016/s0167-4781(01)00359-1. [DOI] [PubMed] [Google Scholar]
- 29.Ebert SN. Rong Q. Boe S. Thompson RP. Grinberg A. Pfeifer K. Targeted insertion of the Cre-recombinase gene at the phenylethanolamine 3-methyltransferase locus: a new model for studying the developmental distribution of adrenergic cells. Dev Dyn. 2004;231:849–858. doi: 10.1002/dvdy.20188. [DOI] [PubMed] [Google Scholar]
- 30.Tompers DM. Labosky PA. Electroporation of murine embryonic stem cells: a step-by-step guide. Stem Cells. 2004;22:243–249. doi: 10.1634/stemcells.22-3-243. [DOI] [PubMed] [Google Scholar]
- 31.Maltsev VA. Rohwedel J. Hescheler J. Wobus AM. Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech Dev. 1993;44:41–50. doi: 10.1016/0925-4773(93)90015-p. [DOI] [PubMed] [Google Scholar]
- 32.Wobus AM. Wallukat G. Hescheler J. Pluripotent mouse embryonic stem cells are able to differentiate into cardiomyocytes expressing chronotropic responses to adrenergic and cholinergic agents and Ca2+ channel blockers. Differentiation. 1991;48:173–182. doi: 10.1111/j.1432-0436.1991.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 33.Carpenedo RL. Sargent CY. McDevitt TC. Rotary suspension culture enhances the efficiency, yield, and homogeneity of embryoid body differentiation. Stem Cells. 2007;25:2224–2234. doi: 10.1634/stemcells.2006-0523. [DOI] [PubMed] [Google Scholar]
- 34.Pertoft H. Fractionation of cells and subcellular particles with Percoll. J Biochem Biophys Methods. 2000;44:1–30. doi: 10.1016/s0165-022x(00)00066-x. [DOI] [PubMed] [Google Scholar]
- 35.Simpson P. Savion S. Differentiation of rat myocytes in single cell cultures with and without proliferating nonmyocardial cells. Cross-striations, ultrastructure, and chronotropic response to isoproterenol. Circ Res. 1982;50:101–116. doi: 10.1161/01.res.50.1.101. [DOI] [PubMed] [Google Scholar]
- 36.Christensen G. Minamisawa S. Gruber PJ. Wang Y. Chien KR. High-efficiency, long-term cardiac expression of foreign genes in living mouse embryos and neonates. Circulation. 2000;101:178–184. doi: 10.1161/01.cir.101.2.178. [DOI] [PubMed] [Google Scholar]
- 37.Ebert SN. Thompson RP. Embryonic epinephrine synthesis in the rat heart before innervation: association with pacemaking and conduction tissue development. Circ Res. 2001;88:117–124. doi: 10.1161/01.res.88.1.117. [DOI] [PubMed] [Google Scholar]
- 38.Linask KK. Han MD. Artman M. Ludwig CA. Sodium-calcium exchanger (NCX-1) and calcium modulation: NCX protein expression patterns and regulation of early heart development. Dev Dyn. 2001;221:249–264. doi: 10.1002/dvdy.1131. [DOI] [PubMed] [Google Scholar]
- 39.Müller JG. Isomatsu Y. Koushik SV. O'Quinn M. Xu L. Kappler CS. Hapke E. Zile MR. Conway SJ. Menick DR. Cardiac-specific expression and hypertrophic upregulation of the feline Na(+)-Ca(2+) exchanger gene H1-promoter in a transgenic mouse model. Circ Res. 2002;90:158–164. doi: 10.1161/hh0202.103231. [DOI] [PubMed] [Google Scholar]
- 40.Cao F. van der Bogt KE. Sadrzadeh A. Xie X. Sheikh AY. Wang H. Connolly AJ. Robbins RC. Wu JC. Spatial and temporal kinetics of teratoma formation from murine embryonic stem cell transplantation. Stem Cells Dev. 2007;16:883–891. doi: 10.1089/scd.2007.0160. [DOI] [PubMed] [Google Scholar]
- 41.Nussbaum J. Minami E. Laflamme MA. Virag JA. Ware CB. Masino A. Muskheli V. Pabon L. Reinecke H. Murry CE. Transplantation of undifferentiated murine embryonic stem cells in the heart: teratoma formation and immune response. FASEB J. 2007;21:1345–1357. doi: 10.1096/fj.06-6769com. [DOI] [PubMed] [Google Scholar]
- 42.Otsu K. Kuruma A. Yanagida E. Shoji S. Inoue T. Hirayama Y. Uematsu H. Hara Y. Kawano S. Na+/K+ ATPase and its functional coupling with Na+/Ca2+ exchanger in mouse embryonic stem cells during differentiation into cardiomyocytes. Cell Calcium. 2005;37:137–151. doi: 10.1016/j.ceca.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Fujimaki K. Aratani Y. Fujisawa S. Motomura S. Okubo T. Koyama H. DNA topoisomerase II inhibitors enhance random integration of transfected vectors into human chromosomes. Somat Cell Mol Genet. 1996;22:279–290. doi: 10.1007/BF02369567. [DOI] [PubMed] [Google Scholar]
- 44.Shaw-White JR. Denko N. Albers L. Doetschman TC. Stringer JR. Expression of the lacZ gene targeted to the HPRT locus in embryonic stem cells and their derivatives. Transgenic Res. 1993;2:1–13. doi: 10.1007/BF01977675. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z. Troilo PJ. Wang X. Griffiths TG. Pacchione SJ. Barnum AB. Harper LB. Pauley CJ. Niu Z. Denisova L. Follmer TT. Rizzuto G. Ciliberto G. Fattori E. Monica NL. Manam S. Ledwith BJ. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11:711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- 46.Subramaniam A. Jones WK. Gulick J. Wert S. Neumann J. Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J Biol Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- 47.Kolossov E. Bostani T. Roell W. Breitbach M. Pillekamp F. Nygren JM. Sasse P. Rubenchik O. Fries JW. Wenzel D. Geisen C. Xia Y. Lu Z. Duan Y. Kettenhofen R. Jovinge S. Bloch W. Bohlen H. Welz A. Hescheler J. Jacobsen SE. Fleischmann BK. Engraftment of engineered ES cell-derived cardiomyocytes but not BM cells restores contractile function to the infarcted myocardium. J Exp Med. 2006;203:2315–2327. doi: 10.1084/jem.20061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swijnenburg RJ. Schrepfer S. Govaert JA. Cao F. Ransohoff K. Sheikh AY. Haddad M. Connolly AJ. Davis MM. Robbins RC. Wu JC. Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts. Proc Natl Acad Sci USA. 2008;105:12991–12996. doi: 10.1073/pnas.0805802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teng CJ. Luo J. Chiu RC. Shum-Tim D. Massive mechanical loss of microspheres with direct intramyocardial injection in the beating heart: implications for cellular cardiomyoplasty. J Thorac Cardiovasc Surg. 2006;132:628–632. doi: 10.1016/j.jtcvs.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 50.Tran N. Li Y. Maskali F. Antunes L. Maureira P. Laurens MH. Marie PY. Karcher G. Groubatch F. Stoltz JF. Villemot JP. Short-term heart retention and distribution of intramyocardial delivered mesenchymal cells within necrotic or intact myocardium. Cell Transplant. 2006;15:351–358. doi: 10.3727/000000006783981918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.