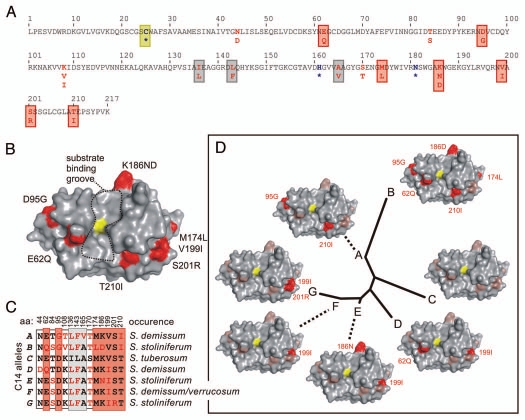
Figure 1.
Structural models of variance in C14 protease domains. (A) Amino acid polymorphism in potato C14 sequences. Only the mature protease domain of S. tuberosum C14 is shown. There are 12 polymorphic amino acids found in wild potato (red), of which seven (boxed red) locate in a ring surrounding the active site and three (boxed grey) locate inside the protein structure. The catalytic residues are indicated in bold blue and with an asterisk (*) and the catalytic Cys is boxed yellow. (B) Model of C14 based on crystal structure of 1S4V. The catalytic Cys is shown in yellow and is in the middle of the substrate binding cleft, which runs from top to bottom. Locations of polymorphic residues are indicated in red and reside in a “ring of fire” around the substrate binding groove. (C) Summary of the occurrence of polymorphic residues in potato C14. There are seven alleles (A–G, left), that differ at 12 positions summarized in the matrix. These were found in wild potato species and cultivated potato (S. tuberosum). (D) Structural polymorphism and phylogenetic relationship of potato C14 proteases. An unrooted tree was generated with the protease domains of C14. The residues indicated in red are different when compared to the protein sequence of cultivated potato (allele C).