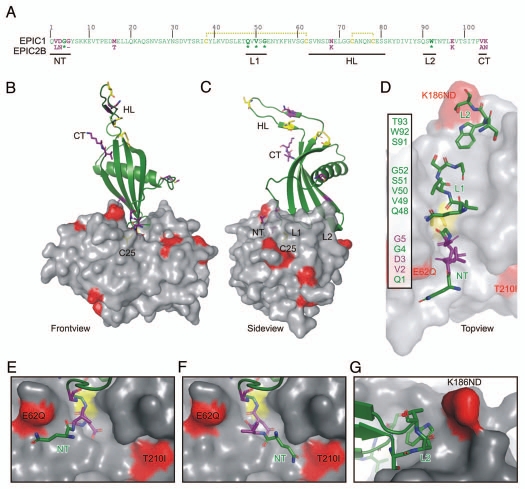
Figure 2.
Model of the EPIC-C14 complex. (A) EPIC 1 protein sequence, without signal peptide. EPIC1 and EPIC2B differ at 8 positions (purple). Residues in the N-terminus (NT), loop-1 (L1) and loop-2 (L2) are directly interacting with the protease. Conserved residues (*) are indicated bold. The hypothetical loop (HT) could not be modelled on the 3IMA template. EPIC1 contains four cysteines (yellow) which are likely connected by disulphide bridges (yellow dashed lines). (C and D) Front-(B), side-(C) and top-(D) views of the model of the EPIC-C14 complex, based on replacement of the tarocystatin-papain crystal structure (3IMA). EPIC 1 is shown as a cartoon (green) and C14 as a semi-transparent surface (gray). The catalytic Cys (C25, yellow) is visible through the transparent surface. As with all cystatins, EPIC s would interact with the substrate binding site of C14 as a wedge consisting of the N-terminus (NT) and two loops (L1 and L2). Polymorphic residues in C14 are indicated in red. Polymorphic sites in EPIC and the cysteins are shown as sticks in purple and yellow, respectively. (D) Model of EPIC residues binding to the substrate binding groove of C14. Residues in the N-terminus (NT) and loops L1 and L2 are shown as sticks. Residues that are polymorphic with EPIC 2B are shown in purple. (E and F) The N-terminus of EPIC may stretch into the left (E) of right (F) cavities of the non-prime substrate binding sites of C14. (G) The L2 loop of EPIC s may interact with the K168ND polymorphic site in C14.