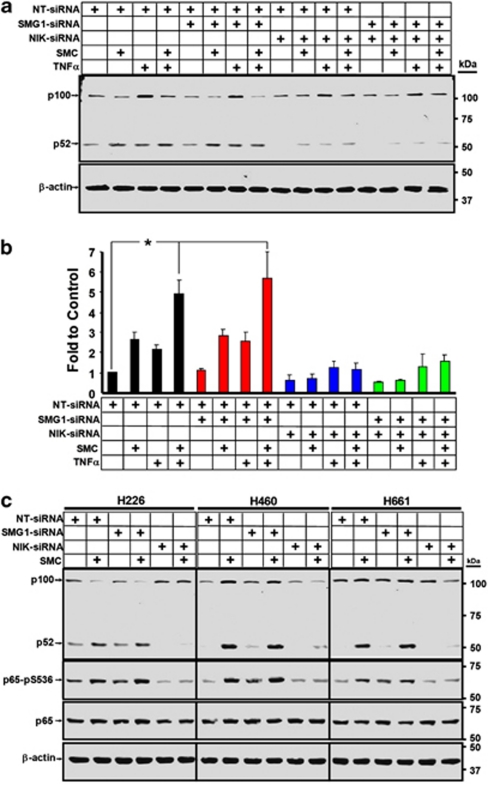
Figure 6.
SMC treatment activates both the classical and alternative NF-κB pathways via NIK, promoting c-FLIP transcription. (a) H226 cells were transfected with siRNA targeting SMG1, NIK or non-targeting siRNA as a control for 24 h, and then exposed to combinations of 100 nM SMC-AEG40730 and 100 ng/ml TNFα for an additional 24 h. Cells were harvested and subjected to western immunoblotting with antibodies recognizing p100 and p52. β-Actin was used as a loading control. (b) H226 cells treated as in (a) were harvested and RNA was isolated for real-time qRT-PCR to determine c-FLIP mRNA levels. Fold changes from four independent experiments relative to non-targeting siRNA and vehicle ±S.E.M. normalized against GAPDH mRNA levels were plotted; siRNA targeting SMG1: red bars; NIK: blue bars; both: green bars; non-targeting siRNA: black bars. Numbers represent the average ±S.E.M. of at least three assays. *P<0.001 significantly different from non-targeting siRNA and vehicle, as determined by ANOVA with post hoc Tukey's HSD. (c) H226, H460 and H460 cells were transfected with siRNA targeting SMG1, NIK or non-targeting siRNA as a control. At 24 h post-transfection, cells were treated with 100 nM SMC-AEG40730 for an additional 24 h. Cells were then harvested and subjected to western immunoblotting with antibodies recognizing p100, p52, phospho-p65 and p65. Downregulation of NIK blocks SMC-induced activation of the classical and alternative NF-κB pathways, as well as SMC- and TNFα-mediated induction of c-FLIP transcripts