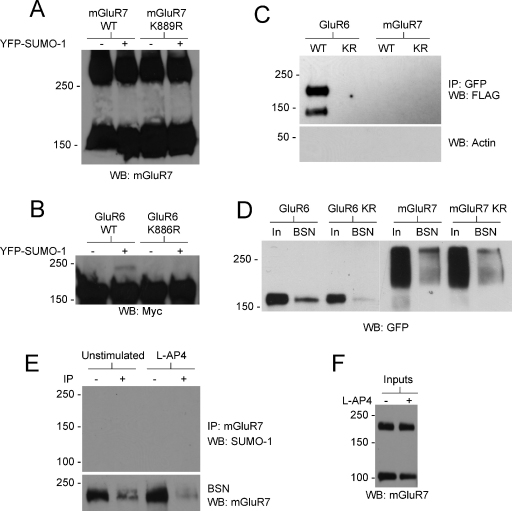
Fig. 4.
SUMOylation of mGluR7 was not detected in HEK293 cells or neurons. (A) SEP-mGluR7 was co-transfected into HEK293 cells in the presence or absence of FLAG-Ubc9 and YFP-SUMO-1. Cells were lysed in SDS-PAGE loading buffer and subjected to Western blotting for mGluR7. The two bands represent monomers and dimers of mGluR7, but a band representing a SUMO-modified form of mGluR7 was not detected. (B) As (A), except cells were transfected with a myc-tagged form of the kainate receptor subunit GluR6 or the non-SUMOylatable K886R mutant. Note the presence of a SUMOylated higher molecular weight form of GluR6 upon expression of YFP-SUMO-1 which is not present for the K886R mutant. (C) HEK293 cells transfected with YFP-GluR6, SEP-mGluR7 or their non-SUMOylatable mutants (denoted as KR) along with FLAG-Ubc9 and FLAG-SUMO-1. GFP-tagged proteins were immunoprecipitated with GFP-trap beads and Western blotted for FLAG. (D) Inputs and equivalent amounts of bead supernatant (BSN) from IPs shown in (C) showing successful depletion (pulldown) of target proteins. (E) 14DIV cortical neurons were lysed, mGluR7 immunoprecipitated and blotted for SUMO-1 (upper panel). Bead supernatant (BSN) was also run alongside an equivalent amount of input to determine pulldown of mGluR7 was successful. (F) Inputs corresponding to the IPs shown in (E).