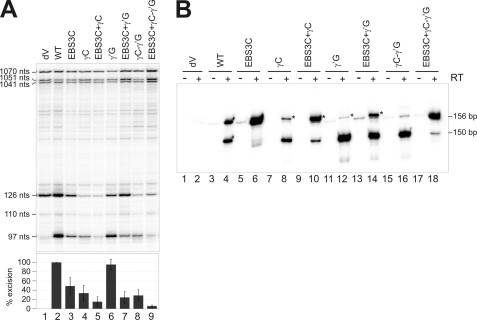
FIGURE 6.
Effect of γ-γ′ mutations on the splicing reaction in vivo. A, total RNA from the S. meliloti RMO17 strain harboring the wild-type intron (pICGterΔORF/pKGIEP), the splicing-defective mutant dV and the corresponding mutants, was reverse-transcribed with an intron-specific primer (P). The cDNA products obtained from excised intron RNA (97 nt) and unspliced precursor derived transcripts (110 and 126 nt) are indicated, together with the more slowly migrating bands corresponding to a characteristic pattern of primary precursor transcripts (1041, 1051, and 1070 nt). Splicing efficiency is plotted as a percentage of wild-type values, with error bars indicating S.D. B, RmInt1-derived products were identified by RT-PCR. PCR was carried out with (+) and without (−) prior RT of RNA from S. meliloti RMO17 harboring the indicated constructs. Total RNA was isolated with the RNeasy Mini Kit (Qiagen), which minimizes the amount of DNA present in the RNA samples. The type of product amplified is indicated (156 and 150 bp, respectively). The products of the mutants marked with an asterisk have an extra cytosine residue at the junction.