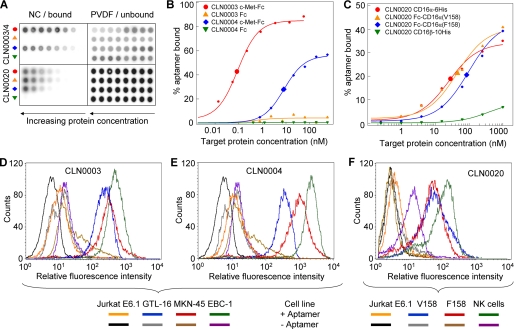
FIGURE 2.
Biochemical and cellular binding characterization of CD16α and c-Met specific aptamers. A, representative dot blot raw data of three DNA aptamers CLN0003, CLN0004, and CLN0020. NC, nitrocellulose membrane readout; PVDF, polyvinylidene difluoride membrane readout. B, fitted binding curves for CLN0003 and CLN0004 to c-Met or Fc. C, fitted CLN0020 binding to both CD16α allotypes and minor binding to CD16β. Enlarged symbols represent calculated KD values. D, FACS analysis of CLN0003 binding to c-Met-positive GTL-16, MKN-45, and EBC-1 cells as compared with c-Met-negative control Jurkat E6.1 cells. E, CLN0004 binding to Met-positive GTL-16, MKN-45, and EBC-1 cells as compared with Jurkat E6.1. F, CLN0020 binding to NK cells and to both alloforms of CD16α presented on recombinant Jurkat cells, in comparison with the parental, CD16-negative Jurkat E6.1 cell line.