Abstract
The helper-component proteinase (HC-Pro) of potyvirus is involved in polyprotein processing, aphid transmission, and suppression of antiviral RNA silencing. There is no high resolution structure reported for any part of HC-Pro, hindering mechanistic understanding of its multiple functions. We have determined the crystal structure of the cysteine protease domain of HC-Pro from turnip mosaic virus at 2.0 Å resolution. As a protease, HC-Pro only cleaves a Gly-Gly dipeptide at its own C terminus. The structure represents a postcleavage state in which the cleaved C terminus remains tightly bound at the active site cleft to prevent trans activity. The structure adopts a compact α/β-fold, which differs from papain-like cysteine proteases and shows weak similarity to nsP2 protease from Venezuelan equine encephalitis alphavirus. Nevertheless, the catalytic cysteine and histidine residues constitute an active site that is highly similar to these in papain-like and nsP2 proteases. HC-Pro recognizes a consensus sequence YXVGG around the cleavage site between the two glycine residues. The structure delineates the sequence specificity at sites P1–P4. Structural modeling and covariation analysis across the Potyviridae family suggest a tryptophan residue accounting for the glycine specificity at site P1′. Moreover, a surface of the protease domain is conserved in potyvirus but not in other genera of the Potyviridae family, likely due to extra functional constrain. The structure provides insight into the catalysis mechanism, cis-acting mode, cleavage site specificity, and other functions of the HC-Pro protease domain.
Keywords: Crystal Structure, Cysteine Protease, Plant Viruses, Protease, Viral Protease, X-ray Crystallography, Autoproteolysis, Potyvirus
Introduction
The genomes of many viruses are translated into polyprotein precursors that are further processed by proteolytic cleavage into mature products (1). Potyviruses are positive-sense single-stranded RNA viruses and form the largest genus of plant virus (2). The genome of potyvirus is translated into a single large 340–370-kDa polyprotein that contains in the N to C order 10 proteins: P1, HC-Pro, P3, 6K1, CI, 6K2, VPg, NIa-Pro, Nlb, and coat protein. The polyprotein is processed into mature products by three virus-encoded proteases P1 (3, 4), helper-component proteinase (HC-Pro)2 (5, 6), and NIa-Pro (7). P1 and HC-Pro cleave only at their respective C termini in cis, whereas NIa-Pro is a trypsin-like cysteine proteinase responsible for the remaining of seven cleavage events in the C-terminal two-thirds of the polyprotein.
HC-Pro is a multifunctional protein associated with polyprotein processing, aphid transmission, and various defense-related functions (2, 8). HC-Pro is generally divided into three functional domains: a N-terminal domain, a central region, and a cysteine protease domain (CPD) in the C-terminal region. HC-Pro was first identified as a helper component for aphid-mediated plant-to-plant transmission of potyvirus (9), and it likely functions as a bridge between the aphid stylet and virions. Mutagenesis studies showed that a KITC motif in the N-terminal domain is critical for binding the aphid stylets (10), and a PTK motif close to the CPD interacts with viral capsid protein (11). The N-terminal domain is essential for aphid transmission but is dispensable for virus viability, infectivity and synergism (12, 13).
HC-Pro is involved in genome amplification, long distance movement, pathogenicity, and viral synergism (13–17). These effects are now generally ascribed to the role of HC-Pro as an RNA silencing suppressor (RSS) (18–21). RNA silencing serves as an antiviral defense system in plants, in which viral double-stranded RNAs are processed by Dicer into small interfering RNAs (siRNAs) that guide Argonaute proteins to cleave viral RNAs (22). The RSS activity of HC-Pro was mainly associated with its central region (13, 14, 21) and was also affected by mutations in the CPD (23, 24). The molecular mechanism of HC-Pro RSS activity remains elusive. Recent studies suggested that HC-Pro suppresses RNA silencing by sequestering siRNA duplex, as many other viral RSSs (25, 26). Consistently, both the siRNA binding and RSS activity of plum pox virus HC-Pro were impaired by mutations in a conserved FRNK motif in the central region (27).
HC-Pro cleaves in cis a Gly-Gly dipeptide at its own C terminus (5, 6). The autoproteolytic activity is exclusively intramolecular as a CPD fragment could not cleave a dead enzyme substrate provided in trans (6). This proteolytic activity is essential for virus viability (28). HC-Pro was classified as a cysteine protease because of a cysteine and histidine residue that are essential for autoproteolysis (29). However, the HC-Pro CPD is not homologous with other cellular and viral cysteine proteases beyond the Potyviridae family.
Despite being a subject of considerable interest for >3 decades, only low resolution structures for HC-Pro have been obtained by means of two-dimensional crystallography and electron microscopy (30, 31). In this study, we determined the crystal structure of HC-Pro CPD from turnip mosaic virus (TuMV) at 2.0 Å resolution. The structure provides important insight into the catalysis mechanism, cis-acting mode, cleavage site specificity, and other functions of the HC-Pro CPD.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
The DNA sequence encoding residues 301–458 of TuMV HC-Pro was PCR-amplified and cloned into an engineered pET28a vector, in which the inserted protein was fused to the C terminus of a His6-SMT3 tag. SMT3 is a yeast small ubiquitin-like modifier protein used to increase protein expression level and solubility (32). The protein was expressed in the Escherichia coli Rosetta(DE3) stain, and protein expression was induced with 0.3 mm isopropyl β-d-1-thiogalactopyranoside and growth at 16 °C. Cells were resuspended in phosphate-buffered saline (PBS) containing 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 2 mm KH2PO4 (pH 7.4) and lysed by sonication. After clarification, the lysate supernatant was loaded onto a HisTrap column. The bound protein was washed with PBS buffer and 50 mm imidazole in PBS buffer and eluted with 500 mm imidazole in PBS buffer. The His6-SMT3 tag was cleaved by ULP1 at 4 °C overnight. The sample was diluted 10-fold with PBS buffer and passed through a HisTrap column to remove the His6-SMT3 tag. The flowthrough was concentrated and further purified in a Superdex-75 column equilibrated in 20 mm HEPES-K (pH 7.6) and 100 mm KCl. Because the protein is prone to reversible aggregation at room temperature, all purification and crystallization steps were performed at 4 °C. The protein was labeled with seleno-methionine by inhibition of the methionine biosynthesis pathway (33) and purified as described above.
Crystallization was performed at 4 °C using the hanging-drop vapor diffusion method by mixing 1 μl of protein solution (∼30 mg/ml in 100 mm KCl, 10 mm DTT, and 20 mm HEPES-K (pH 7.6)) and 1 μl of reservoir solution (1 m (NH4)2SO4, 4% (w/v) PEG3350, and 0.1 m BisTris (pH 5.4)). Se-Met-labeled protein was crystallized in 0.1 m NaH2PO4, 0.1 m KH2PO4, 1.0 m NaCl, and 0.1 m MES (pH 6.0) at 4 °C. Cryoprotection was performed by stepwise incubation in 5, 8, 11, 14, 17, and 20% glycerol made in the respective reservoir solution.
Structure Determination
Diffraction data were collected at the Shanghai Synchrotron Radiation Facility beamline BL17U and processed by HKL2000 (34). The crystals belong to space group I422 with one molecule in the asymmetric unit. The structure was solved by selenium single-wavelength anomalous diffraction phasing in SHARP (35). The model was built in COOT (36) and refined in Refmac (37). The final model contains residues 336–458 and 17 water molecules. The RAMPAGE analysis showed that 95.9% of the residues are in a favorable region, 3.3% in an allowed region, and 0.8% in an outlier region (38). The single outlier residue Ser424 is located in a loop (residues 419–426) that was roughly modeled due to weak electron density. Structural figures were created in PyMOL (39).
RESULTS
Structural Description
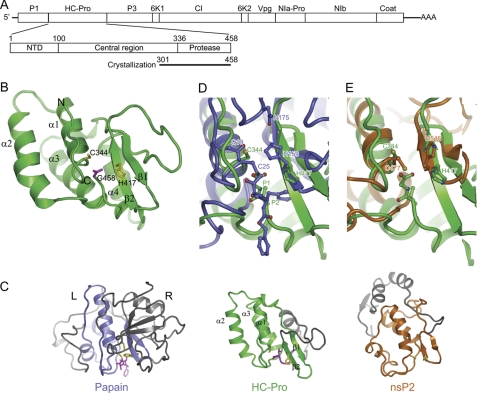
The proteolytic activity of the tobacco etch virus (TEV) HC-Pro was previously shown to reside in 155 residues within its C terminus (5). We expressed, purified, and crystallized a fragment (residues 301–458) that covers a similar C-terminal region of TuMV HC-Pro and corresponds to the mature product of autocleavage (Fig. 1A). The structure was solved by the single-wavelength anomalous diffraction method using a seleno-methionine-labeled crystal and was refined to 2.0 Å with a Rwork/Rfree of 0.246/0.274 (Table 1 and supplemental Fig. S1). The refinement statistics appear worse than what would be expected for a structure with 2.0 Å resolution. The poor refinement statistics may be partially caused by some residues that were not (residues 301–335) or roughly modeled (residues 419–426) because of missing or weak electron density.
FIGURE 1.
Structure of HC-Pro CPD from TuMV. A, diagram of the TuMV RNA genome, the domain organization of HC-Pro, and the construct used for crystallization. Mature products for the polyprotein are indicated. B, ribbon representation of the CPD structure. The secondary structural elements and the N and C terminus are indicated. The catalytic dyad residues Cys344 and His417 (yellow) and the C-terminal residue Gly458 (magenta) are shown as sticks. C, structural comparison of HC-Pro CPD, papain (Protein Data Bank ID code 6PAD), and nsP2 CPD of Venezuelan equine encephalitis alphavirus (Protein Data Bank ID code 2HWK). The three structures are aligned around the catalytic dyad residues. The equivalent structure elements are colored green in HC-Pro, blue in papain, and orange in nsP2, whereas the rest of the structures are gray. The catalytic residues and substrate/inhibitor are shown in sticks and are colored in yellow and magenta, respectively. D and E, superposition of the active site of HC-Pro CPD with that of papain covalently linked to an inhibitor (D) and nsP2 (E).
TABLE 1.
Data collection and refinement statistics
| Crystal form | Native | Se-labeled |
|---|---|---|
| Data collection | ||
| Space group | I422 | I422 |
| Cell dimensions | ||
| a, b, c (Å) | 98.8, 98.8, 65.7 | 99.4, 99.4, 64.8 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.9796 | 0.9796 |
| Resolution range (Å)a | 20–2.00 (2.03–2.00) | 50–2.6 (2.64–2.60) |
| Redundancy | 13.1 (10.6) | 15.4 (13.4) |
| I/σ | 48.7 (4.1) | 67.1 (6.6) |
| Completeness (%) | 98.6 (98.7) | 100 (99.8) |
| Rmergeb | 0.073 (0.528) | 0.057 (0.335) |
| Structure refinement | ||
| Resolution range (Å) | 19–2.0 (2.05–2.0) | |
| No. reflections | 10,612 | |
| No. atoms | 1,008 | |
| Average B factors (Å2) | 56.3 | |
| Rworkc | 0.246 (0.379) | |
| Rfreed | 0.274 (0.351) | |
| Rmsde bond length (Å) | 0.009 | |
| Rmsd bond angles (°) | 1.051 | |
a Values in parentheses are for the data in the highest resolution shell.
b Rmerge = Σ|Ii − Im|/ΣIi, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry related reflections.
c Rwork = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes.
d Rfree is the same as Rwork but calculated on 5% reflections not used in refinement.
e Rmsd, root mean square deviation.
The HC-Pro CPD structure contains residues 336–458 and adopts a compact oval-shaped fold with an overall dimension of 22 Å × 30 Å × 46 Å (Fig. 1B). The secondary structure elements include four α-helices (α1–α4) and two short β-strands (β1 and β2) arranged in the order α1-α2-α3-β1-β2-α4. In addition, two 310 helices are located between α3 and β1 and downstream of α4. The four α-helices form a helix bundle packed against one face of a short β-hairpin formed by strands β1 and β2.
Residues 301–335 were not resolved in the structure, likely due to conformational flexibility and partial degradation. SDS-PAGE analysis of dissolved crystals revealed a mixture of both intact and degraded proteins. This region harbors a PTK motif (residues 310–312 in TuMV) that was shown to probably bind the viral coat protein during aphid-mediated transmission (11).
The catalytic residue Cys344 is located at the N terminus of helix α1, and the other catalytic residue His417 is located on strand β2. The substrate binding cleft is lined by the loop connecting helices α2 and α3 and the N-terminal region of helix α1 on one side and by strand β2 on the other side. The C-terminal tail remains tightly bound at the cleft. The bound product would block the enzyme from accepting new substrate, which explains why HC-Pro possesses only cis-proteolysis activity (6).
Comparison with Other Cysteine Protease Structures
We searched the DALI server and did not find a close structural homolog of the CPD (40). Many papain-like cysteine proteases, such as papain, staphopain, ervatamin, and cathepsin family members, were retrieved with a low z-score of 2.0–3.7 and a root mean square deviation of 3.4–4.0 Å over ∼80 Cα pairs. The overall structure of HC-Pro CPD differs significantly from papain-like-folds (Fig. 1C). Papain-like folds are generally divided into two domains: an L-domain that is mainly helical and harbors the catalytic cysteine and an R-domain with a predominant β-barrel that provides the catalytic histidine (41). The active site cleft is situated at the interface of the two domains. The HC-Pro CPD is much smaller than papain (122 versus 212 residues). Helices α1–α3 of HC-Pro roughly correspond to the L-domain of papain, whereas the equivalents of HC-Pro strands β1 and β2 are part of a much larger and complex β-barrel in the papain R-domain (Fig. 1C).
By visual comparison of known cysteine protease structures, we found another remote structural homolog, the CPD of nsP2 from Venezuelan equine encephalitis alphavirus (42), which was not identified by the DALI search (Fig. 1C). The CPD of HC-Pro and nsP2 are both compact and share similar topology at the α1-α2-α3-β1-β2 region of HC-Pro, although these structural elements are highly different between HC-Pro and nsP2 in terms of orientation and size. In the HC-Pro CPD structure, the region downstream of strand β2 orients the C terminus into the active site for autoproteolysis. No such elements exist in nsP2 CPD, which is a trans-acting protease, and its additional C-terminal region wraps around the structure opposite to the active cleft (Fig. 1C).
Despite different overall structures, HC-Pro, nsP2, and papain-like proteases share a similar active site configuration with the catalytic cysteine residue located at the N terminus of a helix and the catalytic histidine residue in a β-strand (Fig. 1, D and E). In addition, the substrate of HC-Pro and a covalently linked inhibitor of papain occupy a similar position at the active site cleft (Fig. 1D). The active site of most cysteine proteases contains a catalytic triad in which the third residue is an asparagine (Asn175 in papain), aspartate, or glutamate (43, 44). The third residue orients the imidazole ring of the catalytic histidine and perhaps stabilizes its protonated form. The structure shows that HC-Pro apparently lacks the third catalytic residue, which is similar to nsP2 (42).
In papain, the backbone amide of Cys25 and the side chain amide of Gln19 make up an oxyanion hole that stabilizes the oxyanion of the tetrahedral transition intermediate during catalysis (Fig. 1D). No structural equivalence of Gln19 is present in HC-Pro; nevertheless, the amides of Cys344 and Tyr343 might form the oxyanion hole (Fig. 2D).
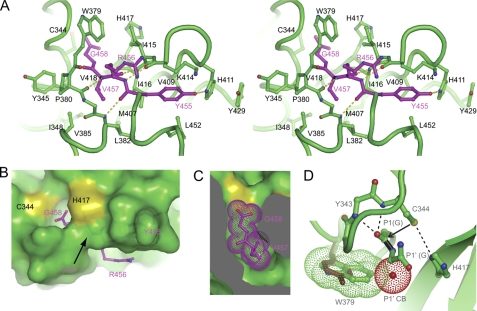
FIGURE 2.
Recognition of the cleavage site. A, Cross-eye stereo view of the C-terminal tetrapeptide (P1–P4) bound at the active site cleft. Interacting residues are shown in stick-and-ball format. Oxygen atoms are red, nitrogen atoms are blue, and carbon atoms are magenta in the tetrapeptide and green for other residues. Dashed lines denote hydrogen bonds. B, surface view of the active site cleft with the bound C-terminal tetrapeptide. C, cut-away view of the S1 and S2 pockets. The P1 residue Gly458 and P2 residue Val457 are shown as dots. D, structural model of the precleavage state. The side chains of the catalytic dyad residues are reoriented to mimic an active configuration, in which Cys344 is deprotonated by His417 and acts as a nucleophile to attack the carbonyl carbon of the scissile bond (black). The amides of Cys344 and Tyr343 may constitute an oxyanion hole to stabilize the transition intermediate. If P1′ has a CB atom (red dots), the CB atom would clash with Trp379 (green dots). Replacement of Trp379 by tyrosine (brown) observed in Bymovirus may remove the specificity for P1′ glycine.
The side chains of the catalytic dyad residues in the HC-Pro CPD structure apparently adopt an inactive conformation; His417 ND1 and Cys344 SG are too distant (7.2 Å) to interact with each other (Fig. 1D). This may be a feature of the postcleavage state or due to a crystal packing interaction involving the His417 imidazole group and the lack of the third catalytic residue stabilizing His417 conformation.
To study the conformation of the enzyme before cleavage, we made constructs that contained mutations C344S or H417K at the catalytic dyad to inactivate the enzyme and the C terminus extended to residue 461. Unfortunately, all of these mutant proteins failed to crystallize. Hence, we resorted to modeling the precleavage state by adding the P1′ residue (Gly459) and orientating the side chains of the catalytic dyad residues (Fig. 2D). According to the classic chemical mechanism of cysteine proteases (43), the catalytic residues Cys344 and His417 likely form a thiolate-imidazolium ion pair. The SG atom of Cys344 would make a nucleophilic attack on the carbonyl carbon of Gly458, resulting in the formation of a thioester-bonded acylenzyme intermediate and release of the amine product. The carboxylate product is released upon hydrolysis of the thioester bond. Our structure corresponds to the postcleavage state with the carboxylate product bound at the active site.
Recognition of the Cleavage Site
Previous mutagenesis analysis demonstrated that HC-Pro recognizes the consensus sequence YXVGG around the cleavage site, in which the cleavage occurs between the two glycine residues (45). According to the standard convention (46), the five residues are named in the N to C order as P4, P3, P2, P1, and P1′, and their respective enzyme binding sites are called S4, S3, S2, S1, and S1′. Our structure delineates the specificity determinants for P4 to P1 (Fig. 2A). The P4–P1 segment adopts an extended structure and sandwiches between strand β2 and the loop connecting helices α1 and α2 by forming four hydrogen bonds along the backbone.
The P1 residue Gly458 is fully encircled by a hole composed of the backbone atoms of Gly342, Tyr343, Cys344, Ile416, His417, and Val418 and the side chain of Trp379 (Fig. 2, B and C). Any side chain at the P1 position would lead to a steric clash with the hole. The P2 residue Val457 is entirely buried, and its side chain is bound at a hydrophobic pocket lined by residues Tyr345, Ile348, Leu382, Val385, Met407, Ile416, and Val418. In addition, the backbone atom of Val457 is shielded from solvent by the side chains of Trp379 and Ile415, which form a bridge across the substrate binding cleft. The side chain of the P3 residue Arg456 is exposed to solvent and not recognized, which is consistent with the sequence variability at this position.
The P4 residue Tyr455 is largely buried at the S4 subsite composed of Val409, His411, Lys414, Ile415, IIe416, Ser451, and Leu452 (Fig. 2A). The phenol ring of Tyr455 is sandwiched between the backbone atoms of Lys414 and Ile415 on one face and the side chain of Leu452 on the other face. One edge of the Tyr455 phenol ring is docked against Val409 and Ile416, whereas the other edge is largely exposed. The hydroxyl group of Tyr455 is specified by a hydrogen bond with the imidazole ring of His411, which further stacks over the side chain of Tyr429. Alanine substitution of the equivalent residues of Asp410 and His411 in TEV HC-Pro (mutant AS 23) was shown to impair autoproteolytic activity (14). The H411A mutation likely disturbs the binding of the P4 tyrosine residue to affect catalytic activity. Interestingly, the HC-Pro of Bymovirus does not display specificity for the P4 tyrosine (Fig. 3, A and B). The S4 subsite residues Val409 and His411 are invariant in HC-Pro homologs that have a P4 specificity; however, in Bymovirus, His411 is replaced by a proline or alanine, and Val409 is replaced by a lysine or phenylalanine. These changes within the S4 subsite may account for the loss of P4 specificity in Bymovirus.
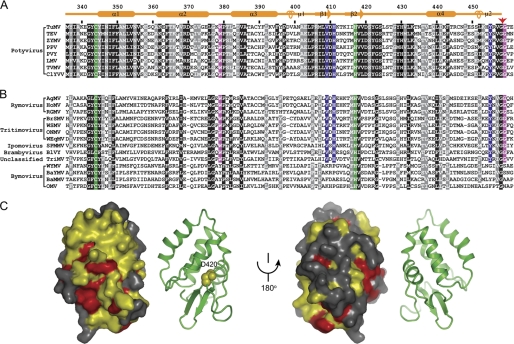
FIGURE 3.
Sequence conservation in the HC-Pro CPD. A, alignment of sequences from the Potyvirus genus. The black and gray shading represent 100 and 90% sequence similarity in 59 aligned sequences, respectively, eight of which are displayed. Similarity groups are defined as follows: S and T, D and E, K and R, and L, I, V, M, F, Y, and W. The secondary structure elements and residue numbers are indicated for TuMV HC-Pro. The red arrow points to the self-cleavage site. The catalytic dyad residues are shaded in green. The P1′ residue Gly458, the putative S1′ residue Trp379 and their equivalents that are invariant in other sequences are shaded in magenta. The P4 residue Tyr455, the S4 residues Val409 and His411 and their equivalents invariant in other sequences are shaded in yellow. ZYMV, zucchini yellow mosaic virus; PPV, plum pox virus; PVY, potato virus Y; LMV, lettuce mosaic virus; TVMV, tobacco vein mottling virus; ClYVV, clover yellow vein virus. B, conservation in the Potyviridae family. Five potyviral HC-Pro proteins (TuMV, TEV, ZYMV, PPV, and PVY, not shown) and all available homologs from other genera of the Potyviridae family are aligned. Residues with 100 and 90% conservation in these sequences are shaded in black and gray, respectively. The genera are indicated. AgMV, agropyron mosaic virus; HoMV, hordeum mosaic virus; RGMV, ryegrass mosaic virus, BrSMV, brome streak mosaic virus; WSMV, wheat streak mosaic virus; ONMV, oat necrotic mottle virus; WEqMV, wheat eqlid mosaic virus; SPMMV, sweet potato mild mottle virus; BlVY, blackberry virus Y; TriMV, Triticum mosaic virus; WYMV, wheat yellow mosaic virus; BaYMV, barley yellow mosaic virus; BaMMV, barley mild mosaic virus; OMV, oat mosaic virus. C, conserved surface at two opposite orientations. Residues at least 90% conserved in the genus Potyvirus as defined in A are colored yellow, and those at least 90% conserved across the Potyviridae family as defined in B are colored red. The corresponding ribbon representations are shown in parallel. Residue Asp420 is shown as spheres.
The structure contains no P1′ residue, and the mechanism of glycine specificity at P1′ remains undetermined. The structure model at the precleavage state suggests that Trp379 is a key determinant for P1′ specificity (Fig. 2D). In support of this proposal, HC-Pro homologs from Bymovirus have no specificity at P1′ and show a parallel replacement of Trp379 by phenylalanine or tyrosine. A smaller six-member ring at the position of Trp379 may relieve the size restriction on P1′ (Fig. 2D).
HC-Pro in the Potyviridae Family
The Potyviridae family currently includes seven genera: Potyvirus, Rymovirus, Tritimovirus, Bymovirus, Ipomovirus, Macluravirus, and Brambyvirus (47, 48). Homologs of potyviral HC-Pro are found in the Rymovirus, Tritimovirus, Bymovirus, and Brambyvirus genera and in one species of Ipomovirus (Fig. 3B). Whether Macluravirus has HC-Pro is unknown because its complete genome is not available. In the genus Ipomovirus, HC-Pro is present in sweet potato mild mottle virus, but it is not present in three other species: cucumber vein yellowing virus (49), squash vein yellowing virus (50), and cassava brown streak virus (51). The taxonomic statue of sweet potato mild mottle virus was thought to be out of date (50). The equivalent genome position of P1/HC-Pro is occupied by two serine proteases P1a and P1b in cucumber vein yellowing virus and squash vein yellowing virus and a single P1 in cassava brown streak virus.
Only HC-Pro proteins from Rymovirus show sequence homology to the full-length Potyvirus HC-Pro, and those from other genera have homology only to the CPD. This suggests that every function of potyviral HC-Pro may not necessarily be conserved in other genera. For example, HC-Pro of wheat streak mosaic virus, which is a tritimovirus, has been shown to be dispensable for systemic infection (52). To provide insight into unique function of HC-Pro CPD in potyvirus, we mapped the residues that are conserved within the Potyvirus genus or across the Potyviridae family onto the CPD structure.
We found that the residues universally conserved in the Potyviridae family primarily constitute the hydrophobic core of the CPD structure or surround the active site, which is consistent with a role in structure maintenance and catalysis. However, within the Potyvirus genus, the CPD shows a much higher degree of conservation with an average pairwise identity of 64% and an average pairwise similarity of 81% (Fig. 3A). Notably, the surface that spans helices α1 and α2, strands β1 and β2, and the loop between β2 and α3 is highly conserved in the Potyvirus genus (Fig. 3C). This is not simply due to a lack of evolutionary divergence because the opposite surface is not conserved (Fig. 3C). The conserved surface is likely involved in functions other than proteolysis that are specific to Potyvirus (and perhaps Rymovirus). Interestingly, asparagine replacement of the equivalent residue of TuMV Asp420, which is located on this surface and shows little structural stabilization role (Fig. 3C), abolished the RSS activity of TEV HC-Pro (23).
DISCUSSION
The HC-Pro CPD performs a single essential proteolytic cleavage to release its C terminus from the rest of the polyprotein. We have determined the first crystal structure of HC-Pro CPD, providing important insight into its unique fold, catalysis mechanism, substrate specificity, and cis-acting mode. The boundary of CPD (residues 336–458) defined by the structure is slightly shorter at its N terminus than previously defined (6) and excludes the PTK motif that is involved in aphid-mediated virus transmission (11). We show that the HC-Pro CPD structure is divergent from known cysteine protease structures while maintaining a conserved catalytic cleft of papain-like proteases. The structure also verifies that the cysteine and histidine residues shown previously to be essential for catalysis indeed form the catalytic dyad at the active site (29).
The structure of HC-Pro CPD in the postcleavage state suggests that trans-activity of HC-Pro would be inhibited by binding of the cleaved product to the active cleft. Similar autoinhibitory structures were observed for the Sindbis virus nucleocapsid protease and the hepatitis C virus NS2 protease (53, 54). Inhibition by the unreleased product appears to be a common mechanism underlying the action of cis-only proteases.
In an attempt to convert the enzyme into a trans-acting protease, we made a fragment (residues 332–447) with the inhibitory C-terminal tail deleted. The truncated protein failed to be purified, suggesting that the C-terminal tail is important for the integrity of the structure. To our knowledge, there is no previous example that an exclusively cis-acting protease can be converted into a trans-protease by removal of the C-terminal tail.
In the current MEROPS protease database (44), HC-Pro proteins constitute a C6 family with unassigned clan status. The nsP2 protease of alphavirus belongs to the C9 family, which is the only member of the newly created clan CN. The structural similarity between HC-Pro and nsP2 CPDs suggests that the HC-Pro C6 family could be assigned to clan CN. Interestingly, both potyvirus and alphavirus are positive-sense RNA viruses and transmitted by insect vectors (aphid for potyvirus and mosquito for alphavirus). The structural similarity between HC-Pro and nsP2 CPD may be a manifestation of common evolutionary origin or a result of convergent evolution. Clan CN may also include p29 and p48 cysteine proteases from Cryphonectria hypoviruses which show short similar sequences with HC-Pro around the catalytic dyad residues and cleavage site (55, 56).
In addition to proteolysis, the CPD of potyvirus has been associated with RSS activity (23, 24). Although the exact role of CPD in RSS activity remains to be defined, an intact CPD structure seems to be important. Many mutations that abolish RSS activity appear to disrupt the CPD structure (23, 24). We identified a surface on the CPD that is conserved in Potyvirus, but not in other genera of Potyviridae that contain a homologous CPD. The surface might contribute to RSS activity by contacting the central region or by making direct role.
Supplementary Material
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility for assistance in data collection and Dr. James Carrington (Oregon State University) for providing TuMV cDNA.
This work was supported by the Chinese Ministry of Science and Technology through projects 973 (2010CB835402) and 863 (2008AA022310) and the Beijing Municipal Government.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 3RNV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HC-Pro
- helper-component proteinase
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- CPD
- cysteine protease domain
- RSS
- RNA silencing suppressor
- TEV
- tobacco etch virus
- TuMV
- turnip mosaic virus.
REFERENCES
- 1. Dougherty W. G., Semler B. L. (1993) Microbiol. Rev. 57, 781–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Urcuqui-Inchima S., Haenni A. L., Bernardi F. (2001) Virus Res. 74, 157–175 [DOI] [PubMed] [Google Scholar]
- 3. Verchot J., Koonin E. V., Carrington J. C. (1991) Virology 185, 527–535 [DOI] [PubMed] [Google Scholar]
- 4. Mavankal G., Rhoads R. E. (1991) Virology 185, 721–731 [DOI] [PubMed] [Google Scholar]
- 5. Carrington J. C., Cary S. M., Parks T. D., Dougherty W. G. (1989) EMBO J. 8, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrington J. C., Freed D. D., Sanders T. C. (1989) J. Virol. 63, 4459–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carrington J. C., Dougherty W. G. (1987) J. Virol. 61, 2540–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maia I. G., Haenni A., Bernardi F. (1996) J. Gen. Virol. 77, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 9. Govier D. A., Kassanis B. (1974) Virology 61, 420–426 [DOI] [PubMed] [Google Scholar]
- 10. Blanc S., Ammar E. D., Garcia-Lampasona S., Dolja V. V., Llave C., Baker J., Pirone T. P. (1998) J. Gen. Virol. 79, 3119–3122 [DOI] [PubMed] [Google Scholar]
- 11. Peng Y. H., Kadoury D., Gal-On A., Huet H., Wang Y., Raccah B. (1998) J. Gen. Virol. 79, 897–904 [DOI] [PubMed] [Google Scholar]
- 12. Dolja V. V., Herndon K. L., Pirone T. P., Carrington J. C. (1993) J. Virol. 67, 5968–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi X. M., Miller H., Verchot J., Carrington J. C., Vance V. B. (1997) Virology 231, 35–42 [DOI] [PubMed] [Google Scholar]
- 14. Kasschau K. D., Cronin S., Carrington J. C. (1997) Virology 228, 251–262 [DOI] [PubMed] [Google Scholar]
- 15. Cronin S., Verchot J., Haldeman-Cahill R., Schaad M. C., Carrington J. C. (1995) Plant Cell 7, 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rojas M. R., Zerbini F. M., Allison R. F., Gilbertson R. L., Lucas W. J. (1997) Virology 237, 283–295 [DOI] [PubMed] [Google Scholar]
- 17. Pruss G., Ge X., Shi X. M., Carrington J. C., Bowman Vance V. (1997) Plant Cell 9, 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kasschau K. D., Carrington J. C. (1998) Cell 95, 461–470 [DOI] [PubMed] [Google Scholar]
- 19. Brigneti G., Voinnet O., Li W. X., Ji L. H., Ding S. W., Baulcombe D. C. (1998) EMBO J. 17, 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Anandalakshmi R., Pruss G. J., Ge X., Marathe R., Mallory A. C., Smith T. H., Vance V. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13079–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kasschau K. D., Carrington J. C. (2001) Virology 285, 71–81 [DOI] [PubMed] [Google Scholar]
- 22. Ding S. W., Voinnet O. (2007) Cell 130, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Torres-Barceló C., Martín S., Daròs J. A., Elena S. F. (2008) Genetics 180, 1039–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varrelmann M., Maiss E., Pilot R., Palkovics L. (2007) J. Gen. Virol. 88, 1005–1015 [DOI] [PubMed] [Google Scholar]
- 25. Mérai Z., Kerényi Z., Kertész S., Magna M., Lakatos L., Silhavy D. (2006) J. Virol. 80, 5747–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lakatos L., Csorba T., Pantaleo V., Chapman E. J., Carrington J. C., Liu Y. P., Dolja V. V., Calvino L. F., López-Moya J. J., Burgyán J. (2006) EMBO J. 25, 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shiboleth Y. M., Haronsky E., Leibman D., Arazi T., Wassenegger M., Whitham S. A., Gaba V., Gal-On A. (2007) J. Virol. 81, 13135–13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kasschau K. D., Carrington J. C. (1995) Virology 209, 268–273 [DOI] [PubMed] [Google Scholar]
- 29. Oh C. S., Carrington J. C. (1989) Virology 173, 692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Plisson C., Drucker M., Blanc S., German-Retana S., Le Gall O., Thomas D., Bron P. (2003) J. Biol. Chem. 278, 23753–23761 [DOI] [PubMed] [Google Scholar]
- 31. Ruiz-Ferrer V., Boskovic J., Alfonso C., Rivas G., Llorca O., López-Abella D., López-Moya J. J. (2005) J. Virol. 79, 3758–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mossessova E., Lima C. D. (2000) Mol. Cell 5, 865–876 [DOI] [PubMed] [Google Scholar]
- 33. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 34. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 35. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 38. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 40. Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 41. Drenth J., Kalk K. H., Swen H. M. (1976) Biochemistry 15, 3731–3738 [DOI] [PubMed] [Google Scholar]
- 42. Russo A. T., White M. A., Watowich S. J. (2006) Structure 14, 1449–1458 [DOI] [PubMed] [Google Scholar]
- 43. Polgar L. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawlings N. D., Woessner J. F. eds) 2nd Ed., pp. 1072–1079, Elsevier/Academic Press, San Diego [Google Scholar]
- 44. Rawlings N. D., Barrett A. J., Bateman A. (2010) Nucleic Acids Res. 38, D227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carrington J. C., Herndon K. L. (1992) Virology 187, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schechter I., Berger A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 47. Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (2005) Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses, pp. 819–841, Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 48. Carstens E. B. (2010) Arch. Virol. 155, 133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Valli A., Martín-Hernández A. M., López-Moya J. J., García J. A. (2006) J. Virol. 80, 10055–10063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li W., Hilf M. E., Webb S. E., Baker C. A., Adkins S. (2008) Virus Res. 135, 213–219 [DOI] [PubMed] [Google Scholar]
- 51. Mbanzibwa D. R., Tian Y., Mukasa S. B., Valkonen J. P. (2009) J. Virol. 83, 6934–6940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stenger D. C., French R., Gildow F. E. (2005) J. Virol. 79, 12077–12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Choi H. K., Tong L., Minor W., Dumas P., Boege U., Rossmann M. G., Wengler G. (1991) Nature 354, 37–43 [DOI] [PubMed] [Google Scholar]
- 54. Lorenz I. C., Marcotrigiano J., Dentzer T. G., Rice C. M. (2006) Nature 442, 831–835 [DOI] [PubMed] [Google Scholar]
- 55. Choi G. H., Pawlyk D. M., Nuss D. L. (1991) Virology 183, 747–752 [DOI] [PubMed] [Google Scholar]
- 56. Shapira R., Nuss D. L. (1991) J. Biol. Chem. 266, 19419–19425 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.