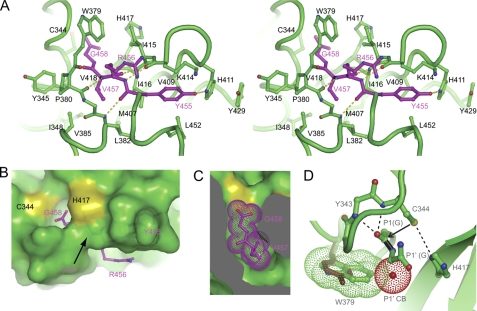
FIGURE 2.
Recognition of the cleavage site. A, Cross-eye stereo view of the C-terminal tetrapeptide (P1–P4) bound at the active site cleft. Interacting residues are shown in stick-and-ball format. Oxygen atoms are red, nitrogen atoms are blue, and carbon atoms are magenta in the tetrapeptide and green for other residues. Dashed lines denote hydrogen bonds. B, surface view of the active site cleft with the bound C-terminal tetrapeptide. C, cut-away view of the S1 and S2 pockets. The P1 residue Gly458 and P2 residue Val457 are shown as dots. D, structural model of the precleavage state. The side chains of the catalytic dyad residues are reoriented to mimic an active configuration, in which Cys344 is deprotonated by His417 and acts as a nucleophile to attack the carbonyl carbon of the scissile bond (black). The amides of Cys344 and Tyr343 may constitute an oxyanion hole to stabilize the transition intermediate. If P1′ has a CB atom (red dots), the CB atom would clash with Trp379 (green dots). Replacement of Trp379 by tyrosine (brown) observed in Bymovirus may remove the specificity for P1′ glycine.