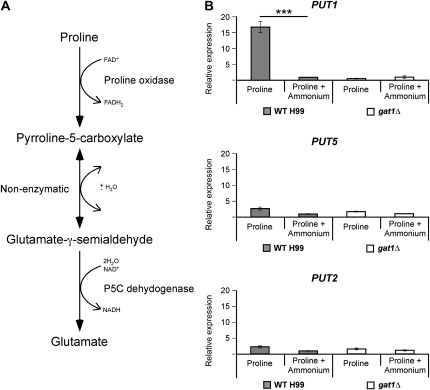
Figure 4.—
Gat1 regulates nitrogen metabolite repression. (A) Scheme representing the predicted proline degradation pathway of C. neoformans. (B) cDNA from wild-type H99 and gat1∆ mutant grown in YNB supplemented with proline or proline plus ammonium (10 mm each nitrogen source) were amplified via qRT-PCR using primers against PUT1 (proline oxidase), PUT5 (proline oxidase), PUT2 (pyrroline-5-carboxylate dehydrogenase), and the control gene ACT1 (actin). One of the predicted catabolic enzyme-encoding genes of proline, PUT1, was sensitive to nitrogen metabolite repression in wild type (*** denotes P < 0.0001) but not in the gat1∆ mutant. The remaining two catabolic genes, PUT5 and PUT2, were nitrogen metabolite repression insensitive in both strains. Error bars represent standard errors across three biological replicates.