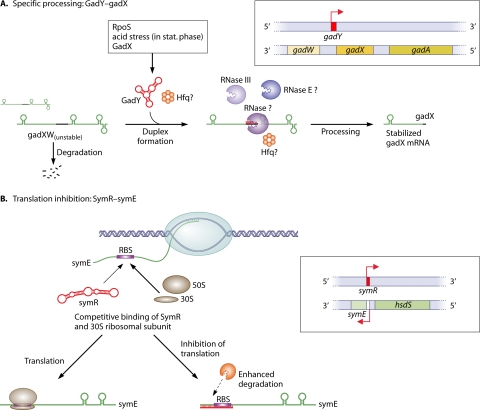
Fig. 3.
Two mechanisms for bacterial asRNAs that overlap at the 3′ or the 5′ end of a protein-coding gene. (A) Specific processing of the gadXW transcript through the asRNA GadY. GadY originates from a gene overlapping the 3′ end of the gadX gene that is highly induced during stationary phase and is dependent on the alternative sigma factor RpoS (66). GadY base pairs with the 3′ untranslated region of the gadX mRNA and confers increased stability through posttranscriptional processing, which allows gadX and gadW mRNA accumulation and increased expression levels of the downstream acid resistance genes (97). Protein factors involved include RNase III, other so-far-unknown RNases (65), probably RNase E (90), and possibly Hfq. (B) Inhibition of translation through SymR (40, 41). SymR is complementary over its full length to the symE 5′ UTR, including the ribosome binding site (RBS), and probably causes a block in ribosome binding and, to a lesser extent, enhanced degradation of the untranslated mRNA. GadY and SymR are drawn according to their RNAfold maximum free energy (mfe) secondary structures.