Abstract
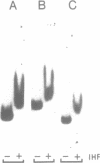
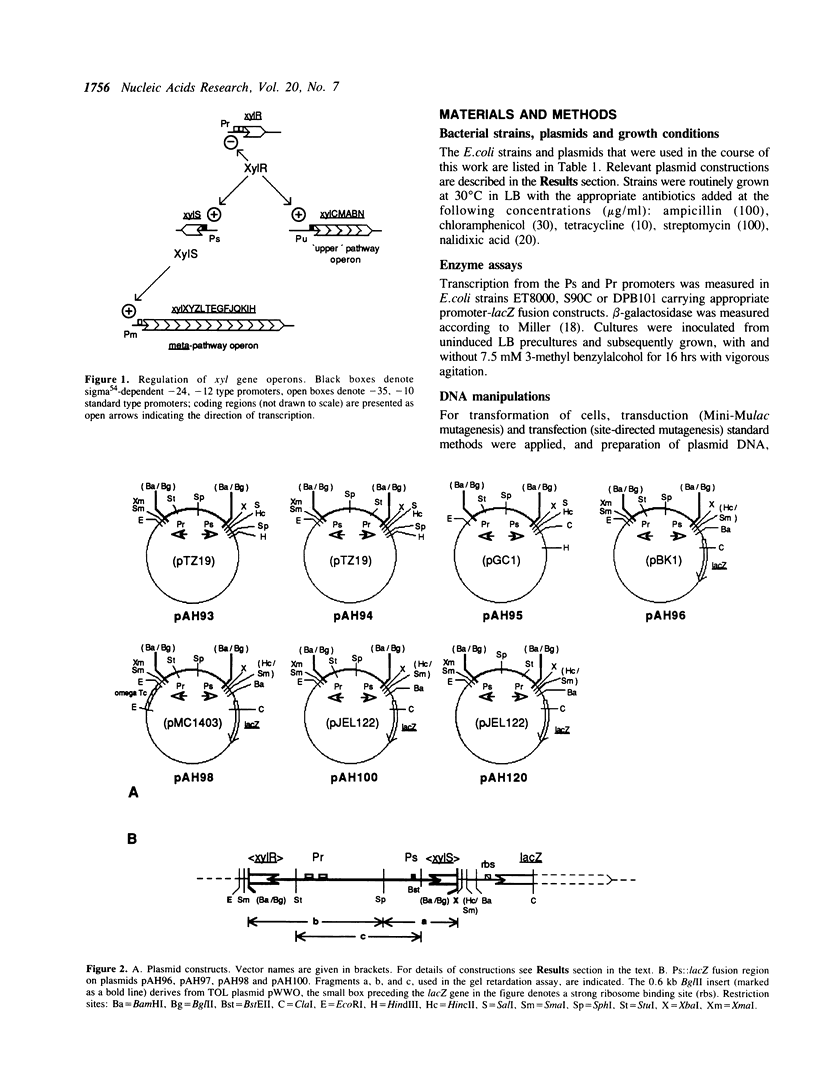
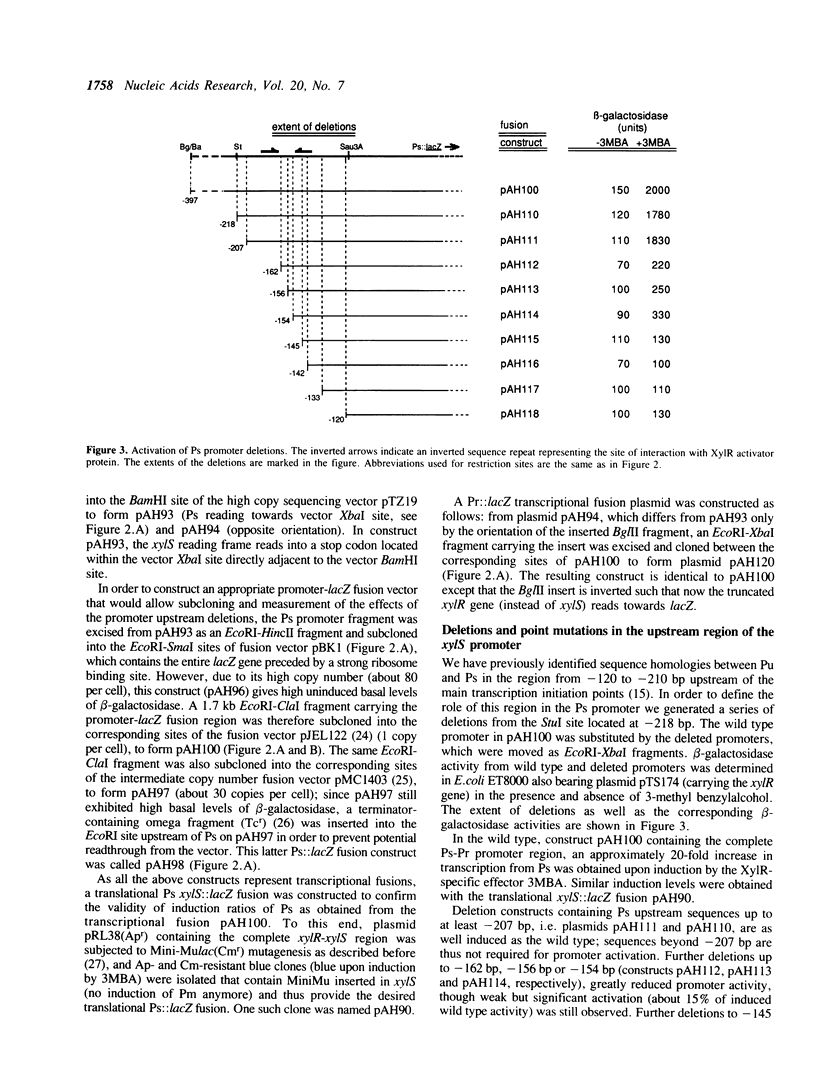
The xylR and xylS genes, which encode the positive regulators of the TOL plasmid catabolic pathways, are adjacent genes on the TOL plasmid and are transcribed from divergent promoters. Transcription from the xylS gene promoter, Ps, is positively regulated by effector-activated XylR protein and requires the specific RNA polymerase sigma 54 subunit (RpoN). Deletions and point mutations in the Ps upstream region localized the site of XylR interaction to the region between -133 bp and -207 bp (with respect to the transcriptional start of the xylS messenger), which contains an inverted sequence repeat largely homologous to the motif recognised by XylR in the XylR-regulated 'upper' catabolic pathway promoter, Pu. Gel retardation experiments showed binding of IHF to the Ps promoter region. Corresponding sequences showing good homology to the IHF-binding consensus were identified close to the Ps Promoter (between -35 bp and -47 bp, Ps proximal site) and further upstream overlapping the XylR recognition sequence (Ps distal site). In the latter case IHF recognition motifs were found well conserved on both strands at nearly the same position (between -140 bp and -152 bp on the upper and between -141 bp and -153 bp on the lower strand). Expression from Ps, either under inducing or non-inducing conditions, was, however, only slightly influenced by the absence of IHF in an IHF-deficient mutant and thus activation of Ps, like that of other sigma 54-dependent promoters which are rich in Ts, does not absolutely require IHF protein.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abril M. A., Buck M., Ramos J. L. Activation of the Pseudomonas TOL plasmid upper pathway operon. Identification of binding sites for the positive regulator XylR and for integration host factor protein. J Biol Chem. 1991 Aug 25;266(24):15832–15838. [PubMed] [Google Scholar]
- Abril M. A., Michan C., Timmis K. N., Ramos J. L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989 Dec;171(12):6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: mutations in the topA gene allow pSC101 replication in the absence of IHF. J Bacteriol. 1989 Apr;171(4):2066–2074. doi: 10.1128/jb.171.4.2066-2074.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon W. V., Kreutzer R., Kent H. M., Morett E., Buck M. Activation of the Klebsiella pneumoniae nifU promoter: identification of multiple and overlapping upstream NifA binding sites. Nucleic Acids Res. 1990 Apr 11;18(7):1693–1701. doi: 10.1093/nar/18.7.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. The xylABC promoter from the Pseudomonas putida TOL plasmid is activated by nitrogen regulatory genes in Escherichia coli. Mol Gen Genet. 1986 Apr;203(1):129–136. doi: 10.1007/BF00330393. [DOI] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Gellert M., Menzel R., Mizuuchi K., O'Dea M. H., Friedman D. I. Regulation of DNA supercoiling in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):763–767. doi: 10.1101/sqb.1983.047.01.087. [DOI] [PubMed] [Google Scholar]
- Gober J. W., Shapiro L. Integration host factor is required for the activation of developmentally regulated genes in Caulobacter. Genes Dev. 1990 Sep;4(9):1494–1504. doi: 10.1101/gad.4.9.1494. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Transcriptional control--lessons from an E. coli promoter data base. Cell. 1991 Aug 9;66(3):415–418. doi: 10.1016/0092-8674(81)90001-5. [DOI] [PubMed] [Google Scholar]
- Hoheisel J., Pohl F. M. Simplified preparation of unidirectional deletion clones. Nucleic Acids Res. 1986 Apr 25;14(8):3605–3605. doi: 10.1093/nar/14.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtel A., Abril M. A., Marques S., Timmis K. N., Ramos J. L. Promoter-upstream activator sequences are required for expression of the xylS gene and upper-pathway operon on the Pseudomonas TOL plasmid. Mol Microbiol. 1990 Sep;4(9):1551–1556. doi: 10.1111/j.1365-2958.1990.tb02066.x. [DOI] [PubMed] [Google Scholar]
- Huang L., Tsui P., Freundlich M. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J Bacteriol. 1990 Sep;172(9):5293–5298. doi: 10.1128/jb.172.9.5293-5298.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Ebina Y., Nakazawa A., Nakazawa T. Nucleotide sequence surrounding transcription initiation site of xylABC operon on TOL plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1688–1691. doi: 10.1073/pnas.81.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Gomada M., Sangodkar U. M., Nakazawa A., Nakazawa T. Upstream regulatory sequence for transcriptional activator XylR in the first operon of xylene metabolism on the TOL plasmid. J Mol Biol. 1990 Nov 20;216(2):251–260. doi: 10.1016/S0022-2836(05)80317-1. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5182–5186. doi: 10.1073/pnas.84.15.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of regulatory gene xylR and operator-promoter regions of the xylABC and xylDEGF operons of the TOL plasmid. J Bacteriol. 1983 Sep;155(3):1192–1199. doi: 10.1128/jb.155.3.1192-1199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T., Harayama S., Ramos J. L., Timmis K. N. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J Bacteriol. 1989 Aug;171(8):4326–4333. doi: 10.1128/jb.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil D. General method, using Mu-Mud1 dilysogens, to determine the direction of transcription of and generate deletions in the glnA region of Escherichia coli. J Bacteriol. 1981 Apr;146(1):260–268. doi: 10.1128/jb.146.1.260-268.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Mermod N., Timmis K. N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987 Nov;1(3):293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- Ratet P., Schell J., de Bruijn F. J. Mini-Mulac transposons with broad-host-range origins of conjugal transfer and replication designed for gene regulation studies in Rhizobiaceae. Gene. 1988;63(1):41–52. doi: 10.1016/0378-1119(88)90544-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santero E., Hoover T., Keener J., Kustu S. In vitro activity of the nitrogen fixation regulatory protein NIFA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7346–7350. doi: 10.1073/pnas.86.19.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui P., Huang L., Freundlich M. Integration host factor binds specifically to multiple sites in the ompB promoter of Escherichia coli and inhibits transcription. J Bacteriol. 1991 Sep;173(18):5800–5807. doi: 10.1128/jb.173.18.5800-5807.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P., Albrechtsen B., Løve Larsen J. E. DNA-protein recognition: demonstration of three genetically separated operator elements that are required for repression of the Escherichia coli deoCABD promoters by the DeoR repressor. EMBO J. 1986 Aug;5(8):2015–2021. doi: 10.1002/j.1460-2075.1986.tb04458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Metzke M., Timmis K. N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the sigma 54-dependent Pu promoter of TOL plasmid. EMBO J. 1991 May;10(5):1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]