Abstract
“Pseudomonas andersonii” is a Gram-negative bacillus initially isolated from a granulomatous lung lesion. Novel species status has not been validated for this single strain. We report four additional cases of pulmonary granuloma involving P. andersonii and further characterize the organism. These patients had pulmonary nodules that were surgically resected and which grew P. andersonii on routine culture. Mycobacterium avium complex was concomitantly isolated in two cases, and fungal structures were identified histopathologically in two other cases. The five P. andersonii strains described to date were similar in growth characteristics, biochemical reactions, matrix-assisted laser desorption ionization–time of flight mass spectrometry protein profiles, and susceptibility to antimicrobial agents. Their 16S rRNA genes were 99.9 to 100% identical but less than 95.0% similar to those of all other known bacteria. The gyrA genes of these strains were 99.5 to 100% identical. These shared features illustrate P. andersonii as a unique and distinct bacterium and support the novel species status of the organism.
INTRODUCTION
Infectious pulmonary granulomas are frequently encountered in pathology practice. They are usually caused by acid-fast bacilli (AFB) or fungi. Gram-negative bacteria are rarely implicated in these cases. Brucella suis (11), Bartonella spp. (7), and Burkholderia cepacia (1) have been isolated from pulmonary granulomas, but they are much more commonly isolated from other clinical sources.
In 2001, a novel species, “Pseudomonas andersonii,” was implicated as the cause of a pulmonary granuloma (6). Nearly a decade later, this remains the only reported case, and the species name has yet to be validated (4). Based on a single case report, the clinical significance of the novel species is unclear. In this study we report four additional cases of lung granulomas also involving P. andersonii, which was identified by gene sequencing. Characterization of these four P. andersonii strains and the previously described strain suggest that they form a distinct, clinically relevant group of bacteria that differs significantly from other known bacterial species.
Case Reports
Case 1.
A 57-year-old male from Connecticut with a history of hepatitis C and peripheral vascular disease was found to have a left lung nodule on a chest X-ray during a routine physical examination. He reported a chronic cough but no shortness of breath. He reported frequent travel to Arizona. A computed tomography (CT) scan confirmed a 1.8-cm contrast-enhancing solitary nodule in the left lower lobe, with peripheral calcifications. A wedge resection was performed. Histopathology demonstrated a necrotizing granulomatous nodule with Coccidioides spherules by hematoxylin and eosin (H&E), Gomori methenamine silver (GMS), and periodic acid-Schiff (PAS) stains. A Ziehl-Neelsen stain was negative for AFB. A tissue Gram stain did not show definitive bacteria. Cultures were negative for AFB and fungi, but a Gram-negative rod was isolated and later identified as P. andersonii by 16S rRNA gene sequencing at ARUP. The patient was not treated with antibiotics because the lesion was excised completely. He was lost to follow-up after discharge from the hospital.
Case 2.
A 77-year-old female from Wisconsin with a history of breast carcinoma and mastectomy 19 years prior presented with right hip pain. Magnetic resonance imaging (MRI) and a positron emission tomography (PET)-CT scan suggested cancer metastases to the hipbones. The PET-CT also revealed a 2-cm lung nodule in the right lower lobe in the costophrenic sulcus, which had increased metabolic activity suggestive of malignancy. The patient had no history of tobacco use and reported no shortness of breath or constitutional symptoms. A video-assisted thoracoscopy with wedge resection was performed. Histopathology demonstrated bronchocentric and interstitial granulomas but no evidence of malignancy. The tissue was negative for AFB and fungi by special stains. A tissue Gram stain, performed later, was inconclusive for bacteria. Cultures grew Mycobacterium avium complex, a few Bacillus sp. colonies, and a Gram-negative bacillus identified as P. andersonii by 16S rRNA sequencing. The patient did well after surgical resection but presented with a pleural effusion 1 year later.
Case 3.
A 58-year-old female from Illinois with a smoking history presented with right-side chest discomfort. A right middle-lobe lung nodule was found on chest X-ray, for which she was followed for 8 months. Because the nodule was enlarging and moderately PET avid, it was surgically resected. An intraoperative frozen section demonstrated a nonnecrotizing granulomatous inflammation associated with polarizable foreign material within giant cells. Routine histopathology confirmed these findings, but AFB, Gram, and GMS stains were negative. P. andersonii, but no AFB or fungi, was isolated from cultures. Upon further sectioning of the tissue, septated fungal hyphae were noted on a PAS stain. However, negative fungal culture precluded a definitive identification. The patient was not treated with antibiotics and was doing well at a visit 2 months postoperatively.
Case 4.
A 49-year-old female from Maryland had been followed for 4 years after presenting with bronchiectasis and hemoptysis. She initially complained of chronic cold and flu symptoms with congestion and a productive cough that was occasionally blood tinged. She had no history of smoking. A chest CT scan revealed a small cavity in the right upper lobe. She was given antibiotics and followed by annual chest CT scans. Three years later an increase in the cavity size was noted along with a soft nodular density adjacent to the cavity wall. A follow-up CT scan showed enlargement of the cavity and of the nodular tissue mass, prompting surgical resection of the mass. Histopathologic examination showed necrotizing granulomatous inflammation. AFB stains demonstrated acid-fast bacilli, but fungal and Gram stains were negative. Cultures grew M. avium complex and a Gram-negative rod later identified as P. andersonii. The patient was treated for M. avium complex with rifampin and ethambutol. She was doing well without respiratory symptoms 4 months postoperatively.
MATERIALS AND METHODS
We reviewed data from more than 20,000 clinical isolates identified at ARUP Laboratories since January 2005 by partial 16S rRNA gene sequencing using Clinical and Laboratory Standards Institute (CLSI) interpretive criteria and the SmartGene IDNS bacterial 16S rRNA gene database and web software (SmartGene, Lausanne, Switzerland) (2). Among them, four isolates showed 99.7 to 100% identity to P. andersonii (ATCC BAA-267 = NCTC 13293; GenBank accession AF291818) over the first 500 bp of the 16S rRNA gene. The four isolates had been stored in brain heart infusion medium (Hardy Diagnostics, Santa Maria, CA) supplemented with 15% glycerol at −70°C since initial laboratory testing. The P. andersonii type strain (ATCC BAA-267) was obtained from the American Type Culture Collection (ATCC).
The P. andersonii strains and control strains of Pseudomonas aeruginosa ATCC 27853T and Pseudomonas stutzeri ATCC 17588T were subcultured on sheep blood agar (SBA; Hardy Diagnostics) for further characterization. Phenotypic tests were performed using conventional reagents and tubed media and included growth at 42°C in tryptic soy broth, growth on cetrimide agar, citrate utilization, pigment production on Tech agar, and reactions for oxidase, catalase, bile-esculin hydrolysis, and urea hydrolysis. Further biochemical and assimilation tests were performed on API NE test strips (bioMérieux, Marcy l'Etoile, France). Susceptibility to 14 antimicrobial agents was determined by broth microdilution using the recommended CLSI guidelines (3).
The protein profile spectra of the study strains were generated using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) on a Bruker Daltonics microflex LT instrument (Billerica, MA). Cellular proteins were extracted by formic acid-acetonitrile. Spectra were generated in the automatic mode using standard MALDI-TOF MS settings and the MALDI Biotyper automation control software (version 2.0; Bruker Daltonics). A reference spectrum (MSP) was created using the P. andersonii type strain. Spectra were compared to the Bruker database of 3,207 bacterial MSP, of which 140 originated from known Pseudomonas spp. Species-level identification was achieved if a matching score was ≥1.9 as described by Seng et al. (8). An MSP dendrogram was generated using the MALDI Biotyper software.
Molecular characterization was performed by sequencing the complete 16S rRNA and partial gyrA genes as previously described (5, 9) and by a BLAST search of the GenBank nucleotide database. The type strain gyrA gene sequence was available as GenBank accession number DQ989183. Sequence alignment and phylogenetic trees were constructed using the neighbor-joining method with Kimura's two-parameter distance correction model and 1,000 bootstrap replications in the MEGA, version 4, software package (10). Selected Pseudomonas reference sequences were obtained from GenBank for phylogenetic comparison.
Patient information, including geographical source, clinical site, age, sex, and associated or underlying disease or risk factors, was compiled under an institutional review board-approved protocol.
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under the accession numbers HM581510 to HM581519 for the ARUP strains.
RESULTS
Characterization of P. andersonii strains.
The clinicopathologic features of these cases and the type strain of P. andersonii are summarized in Table 1. All five isolates grew after overnight incubation at 35°C on SBA as small, smooth, opaque, and light-brown colonies. They were positive for oxidase, catalase, and urease and utilized malic acid. All were negative for bile-esculin hydrolysis and citrate utilization and failed to assimilate most substrates tested. The isolates did not grow on cetrimide agar or at 42°C in tryptic soy broth and did not produce pigment on Tech agar (Table 2). The isolates were susceptible to most antibiotics tested, including ticarcillin-clavulanate, amikacin, imipenem, and ciprofloxacin but resistant to chloramphenicol and minocycline. The isolates were also susceptible to trimethoprim-sulfamethoxazole, in contrast to the first described case (Table 3).
Table 1.
Case histories of patients with Pseudomonas andersonii isolated from pulmonary granulomas
| Case no. | Sexa | Age (yr) | Isolate no. | State | Lesion, pathology | Culture date | Copathogen(s) | Antimicrobial treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 57 | ARUPPA1 | CT | Lung nodule, necrotizing granulomas | February 2008 | Coccidioides immitis | None | Lost to follow-up |
| 2 | F | 77 | ARUPPA2 | WI | Lung nodule, necrotizing granulomas | May 2008 | Mycobacterium avium complex and few Bacillus colonies | None | Recovered |
| 3 | F | 58 | ARUPPA3 | IL | Lung nodule, necrotizing granulomas | November 2008 | Fungus with septate hyphae and foreign body deposition | None | Recovered |
| 4 | F | 49 | ARUPPA4 | MD | Lung nodule, necrotizing granulomas | October 2009 | Mycobacterium avium complex | Rifampin and ethambutol | Recovered |
M, male; F, female.
Table 2.
Biochemical characteristics of Pseudomonas andersonii strains
| Phenotypic testa | P. andersonii BAA-267T | ARUPPA1 | ARUPPA2 | ARUPPA3 | ARUPPA4 | P. aeruginosa ATCC 27853 | P. stutzeri ATCC 17588 |
|---|---|---|---|---|---|---|---|
| Growth | |||||||
| 42°C in TSB broth | − | − | − | − | − | + | + |
| Cetrimide agar | − | − | − | − | − | + | − |
| Pigment on Tech agar | − | − | − | − | − | + | − |
| Oxidase | + | + | + | + | + | + | + |
| Catalase | + | + | + | + | + | + | + |
| Simmons citrate | − | − | − | − | − | + | + |
| Nitrate reduction | − | − | + | + | − | + | + |
| Indole | − | − | − | − | − | − | − |
| Glucose fermentation | − | − | − | − | − | − | − |
| Arginine dihydrolase | − | − | − | − | − | + | − |
| Hydrolysis | |||||||
| Esculin | − | − | − | − | − | − | − |
| Gelatin | − | − | − | − | − | + | − |
| Urea | + | + | + | + | + | + | − |
| Assimilation | |||||||
| d-Glucose | − | − | − | − | − | + | + |
| l-Arabinose | − | − | − | − | − | − | − |
| d-Mannose | − | − | − | − | − | − | − |
| d-Mannitol | − | − | − | − | − | + | − |
| N-Acetylglucosamine | − | − | − | − | − | + | − |
| d-Maltose | − | − | − | − | − | − | + |
| Potassium gluconate | − | − | − | − | − | + | + |
| Capric acid | + | + | + | − | + | + | + |
| Adipic acid | − | − | − | − | − | + | − |
| Malic acid | + | + | + | + | + | + | + |
| Trisodium citrate | − | − | − | − | − | + | + |
| Phenylacetic acid | − | − | − | − | − | − | − |
TSB, tryptic soy broth.
Table 3.
Antimicrobial susceptibilities of Pseudomonas andersonii isolates
| Antibiotic | MIC (μg/ml) |
Interpretationa | ||||
|---|---|---|---|---|---|---|
| BAA-267T | ARUPPA1 | ARUPPA2 | ARUPPA3 | ARUPPA4 | ||
| Amikacin | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | S |
| Gentamicin | <0.25 | <0.25 | <0.25 | <0.25 | <0.25 | S |
| Tobramycin | <2.5 | <2.5 | <2.5 | <2.5 | <2.5 | S |
| Chloramphenicol | 32.0 | 16.0 | 16.0 | 16.0 | 16.0 | I/R |
| Minocycline | 8.0 | 4.0 | 4.0 | 4.0 | 8.0 | I |
| Imipenem | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | S |
| Meropenem | 2.0 | <0.25 | <0.25 | <0.25 | <0.25 | S |
| Ciprofloxacin | <0.12 | <0.12 | <0.12 | <0.12 | <0.12 | S |
| Levofloxacin | <0.25 | <0.25 | <0.25 | 0.5 | <0.25 | S |
| Trimethoprim-sulfamethoxazole | 4.0/76 | 0.5/9.5 | 0.5/9.5 | 0.5/9.5 | 0.25/4.8 | R/S |
| Piperacillin-tazobactam | 2.0/4.0 | <1.0/4.0 | 4.0/4.0 | 2.0/4.0 | 4.0/4.0 | S |
| Ticarcillin-clavulanic acid | <4.0/2.0 | <4.0/2.0 | <4.0/2.0 | <4.0/2.0 | <4.0/2.0 | S |
| Aztreonam | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | S |
| Ceftazidime | 1 | <0.5 | 1 | <0.5 | 1 | S |
Interpretations based on Clinical and Laboratory Standards Institute MIC breakpoints for other non-Enterobacteriaceae; S, susceptible; I, intermediate; R, resistant.
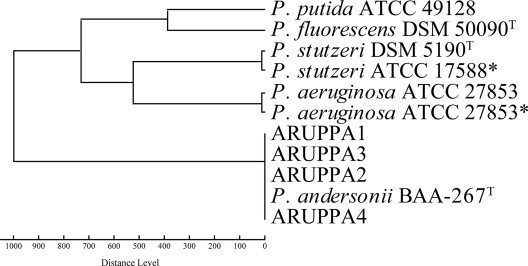
MALDI-TOF MS analysis found no significant similarity to any known species in the default Bruker database (matching scores of <1.4). Control spectra generated from P. stutzeri and P. aeruginosa were identified as their respective species (matching score of >2.3). Upon addition of an MSP of the P. andersonii type strain to the MALDI-TOF database, all four clinical strains achieved species-level identification (matching scores of >2.3). An MSP dendrogram shows the relative similarity of P. andersonii MALDI-TOF spectra compared to other related Pseudomonas spp. (Fig. 1).
Fig. 1.
Dendrogram based on MSP of Pseudomonas andersonii strains and other Pseudomonas references. Asterisk (*) indicates reference MSP generated in-house; all other references were obtained from the Bruker database.
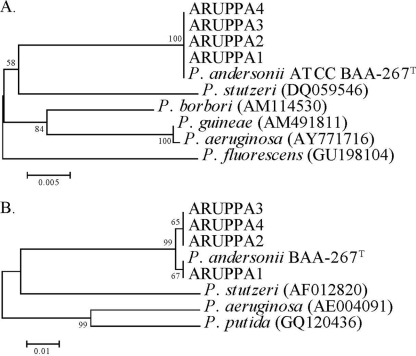
Among the five strains, the full-length 16S rRNA gene sequences shared 99.9 to 100% identity, and the partial gyrA gene sequences were also 99.5 to 100% identical. The next closest species by percent identity in the GenBank database was P. stutzeri for both 16S rRNA and gyrA genes, with its 16S gene (DQ059546) sharing 95.7% identity (1,434 of 1,498 bp) and its gyrA gene (AF012820) sharing 91.1% identity (431 of 473 bp). Other Pseudomonas species, including P. aeruginosa and P. mendocina, showed comparable levels of identity (∼95%) to P. andersonii. Phylogenetic trees based on these gene sequences (Fig. 2) demonstrated species-level phylogenetic distance between P. andersonii and other Pseudomonas species.
Fig. 2.
Neighbor-joining phylogenetic trees of 16S rRNA (A) and gyrA (B) genes of Pseudomonas andersonii strains and other Pseudomonas references. Sequences obtained from GenBank have accession numbers recorded in parentheses. Branch support is recorded at nodes as a percentage of 1,000 bootstrap iterations.
DISCUSSION
The most interesting clinical characteristic of P. andersonii is its association with lung granulomas. Among >20,000 other 16S rRNA sequencing identifications performed at ARUP, P. andersonii has not been identified from specimens other than lung granulomas, suggesting it has a specific clinical niche. In the original case of P. andersonii, the pathogenic role of the bacterium was evident, as it was found to be intimately involved with the necrotic granulomatous lesion and no other organisms could be isolated or demonstrated (6). In the present cases the pathogenic role of P. andersonii is unclear because either a fungus or M. avium complex was also demonstrated or isolated in each case and all patients recovered without the use of antibiotics. That P. andersonii was a laboratory contaminant is unlikely, as one would predict that a contaminant could be isolated from a variety of clinical specimens, not solely pulmonary granulomas. Also, we have described its recovery in different regions of the United States at different times, further arguing against contamination.
P. andersonii is apparently a rare clinical isolate, and its reservoir remains unknown. We speculate an environmental origin because the cocultivated fungi and M. avium complex in the present cases, as well as other known Pseudomonas spp., are environmental organisms. Inhalation of aerosolized organisms may be the most likely route of infection, as is thought to be the case for many fungal and M. avium complex infections. It is remarkable that P. andersonii is able to remain viable in a granulomatous lesion that includes dense mixed components of inflammatory cells and tissue factors. Pre-existing lung injuries, such as those caused by cigarette smoking or bronchiectasis, or developing granulomas by copathogens may favor the involvement of the bacterium. Further studies are required to determine the pathogenic potential and the mechanisms by which it evades the immune defenses.
These cases illustrate that P. andersonii may have an important role as a coinfector, and its identification should be an indication to look for other pathogens. Because tissue specimens are not routinely stained by Gram stain in the course of histopathologic assessment of granulomas, an organism such as P. andersonii may easily be missed. However, it should be noted that we were unable to definitively identify Gram-negative bacilli on a tissue Gram stain of these cases, unlike with the original case described. The relatively slow initial growth rate and the absence of profiles in most identification system databases likely lead to underrecognition of this organism. Although these isolates tested as susceptible to many antimicrobial agents, the available follow-up data of these cases suggest that treatment is not always necessary. Resection of the pulmonary lesion and treatment of the other pathogens identified may be sufficient for patient management, but the number of cases described is small and the follow-up data were limited.
Our genotypic and phenotypic characterizations of the five P. andersonii strains suggest that they form a group of organisms that differ at the species level from other Pseudomonas spp. This conclusion supports the initial proposal of a novel species status. Therefore, we emend the description of the organism as follows.
Description (emended) of Pseudomonas andersonii.
P. andersonii (andersonii, after the institution name M. D. Anderson Cancer Center, the place of its initial isolation) is a Gram-negative bacillus. It measures 0.5 by 0.9 to 2 μm in size and bears a single polar flagellum. The bacterium grows readily in 24 h on several culture media upon subculture, although initial isolation may take longer. It produces catalase and oxidase and hydrolyzes urea. It is asaccharolytic, does not use citrate as a carbon source or hydrolyze bile esculin, and assimilates d-glucose but does assimilate malic acid. The cellular fatty acids consist mainly of straight-chain saturated and polyunsaturated or cyclopropane acids. The organism is susceptible to most antimicrobial agents. So far, this organism has been described only from human nodular lung lesions (granulomas). The type strain is ATCC BAA-267T (= NCTC 13293T), and it was isolated in 2000 in Houston, TX.
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1. Belchis D. A., Simpson E., Colby T. 2000. Histopathologic features of Burkholderia cepacia pneumonia in patients without cystic fibrosis. Mod. Pathol. 13:369–372 [DOI] [PubMed] [Google Scholar]
- 2. CLSI 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI document MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 3. CLSI 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Euzéby J. P. 1997. List of bacterial names with standing in nomenclature: a folder available on the Internet. Int. J. Syst. Bacteriol. 47:590–592 [DOI] [PubMed] [Google Scholar]
- 5. Han X. Y., Andrade R. A. 2005. Brevundimonas diminuta infections and its resistance to fluoroquinolones. J. Antimicrob. Chemother. 55:853–859 [DOI] [PubMed] [Google Scholar]
- 6. Han X. Y., et al. 2001. Pulmonary granuloma caused by Pseudomonas andersonii sp nov. Am. J. Clin. Pathol. 116:347–353 [DOI] [PubMed] [Google Scholar]
- 7. Maguiña C., Gotuzzo E. 2000. Bartonellosis. New and old. Infect. Dis. Clin. North Am. 14:1–22, vii [DOI] [PubMed] [Google Scholar]
- 8. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 9. Simmon K. E., Low Y. Y., Brown-Elliott B. A., Wallace R. J., Jr., Petti C. A. 2009. Phylogenetic analysis of Mycobacterium aurum and Mycobacterium neoaurum with redescription of M. aurum culture collection strains. Int. J. Syst. Evol. Microbiol. 59:1371–1375 [DOI] [PubMed] [Google Scholar]
- 10. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 11. Webb W. A., Thoroughman J. C. 1966. Solitary pulmonary nodule due to Brucella suis: report of a case. Dis. Chest 49:222–224 [DOI] [PubMed] [Google Scholar]