Abstract
The emergence of Scedosporium infections in diverse groups of individuals, which are often treatment refractory, warrants timely and accurate laboratory diagnosis. Species- or group-specific primers based on internal transcribed spacer (ITS) sequence polymorphisms were designed for Scedosporium aurantiacum, Scedosporium dehoogii, Scedosporium prolificans, Pseudallescheria boydii species complex (former clade 5)/Pseudallescheria apiosperma (formerly classified as S. apiospermum sensu lato) and Pseudallescheria minutispora. Primers for S. aurantiacum, S. prolificans, and P. boydii species complex/P. apiosperma were incorporated into a multiplex PCR assay for the detection and identification of the three major clinically important Scedosporium species and validated using sputum specimens collected from patients seen at a major Australian cystic fibrosis clinic. The multiplex PCR assay showed 100% specificity in identifying the three major clinically relevant Scedosporium species from pure culture. When evaluated using DNA extracts from sputa, sensitivity and specificity of the multiplex PCR assay were 62.1% and 97.2%, respectively. This highly species-specific multiplex PCR assay offers a rapid and simple method of detection of the most clinically important Scedosporium species in respiratory tract specimens.
INTRODUCTION
Scedosporium species are emerging fungal pathogens capable of causing a wide range of infections, particularly in seriously ill and immunocompromised patients (8, 26). Long-term colonization with these fungi has been reported in patients with structurally abnormal respiratory airways, including those with cystic fibrosis (CF) (1, 2, 6, 9). Invasive fungal infection is rare in CF patients prior to lung transplantation (15), yet Scedosporium colonization may be a risk factor for invasive infection posttransplantation. Since Scedosporium spp. are resistant to many antifungal agents, colonization is a relative contraindication for transplantation in some centers (8, 24).
Among clinical isolates, the most frequently encountered Scedosporium species include Scedosporium prolificans, Scedosporium apiospermum/Pseudallescheria apiosperma, Pseudallescheria boydii sensu stricto, and the recently described species Scedosporium aurantiacum (11, 13, 14). The emergence of Scedosporium infections in Australia has been highlighted in earlier reports describing the species distribution, clinical epidemiology, and outcomes (1, 2, 7, 10, 16). Further, a nationwide population-based study uncovered a substantial number of infections due to S. aurantiacum and identified an association between isolation of this species and the presence of chronic lung disease (16). Invasive fungal infection is associated with significant morbidity and mortality rates of up to 75 to 80% (17, 25, 26). Therefore, early and accurate detection of Scedosporium infections is crucial for prompt and effective treatment.
Conventional mycological methods for detection and identification of Scedosporium spp. in clinical specimens, however, are insensitive and time-consuming, as culture from sputum samples may require up to 14 days to produce fungal growth adequate for morphological identification (1, 5). A number of molecular detection and identification methods for Pseudallescheria/Scedosporium have been reported (3, 4, 19, 20, 21, 29). Real-time PCR protocols for detection of S. prolificans and S. apiospermum sensu lato have been developed (4). Bouchara et al. employed an oligonucleotide array targeting the internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA) gene cluster for direct detection of a broad range (n = 20 species) of fungal pathogens, including the Pseudallescheria/Scedosporium species complex and S. prolificans in the sputum of CF patients (3). An ITS-directed pan-fungal PCR assay combined with DNA sequencing was used to detect multiple fungal genera, including S. prolificans, in fresh and formalin-fixed clinical specimens (19). Recently, multiplex-tandem PCR (MT-PCR)- and rolling circle amplification (RCA)-based approaches successfully identified S. prolificans and S. apiospermum sensu lato from isolated colonies (both methods) and from blood cultures (MT-PCR) (20, 21, 29). However, most of these assays do not take into account the taxonomic reclassification of Scedosporium spp. or are designed to detect only a few Scedosporium species. Further, they required either an additional sequencing step, which may pose a problem in the case of mixed infections, or need specialized equipment, which may prevent them from being easily implemented into the routine microbiology laboratory in the near future.
In the present study, species-specific primers for the most common Scedosporium species (S. aurantiacum, S. prolificans, and P. boydii species complex/P. apiosperma) were designed, and a multiplex PCR method was developed to enable simultaneous rapid and accurate detection and identification of the major pathogenic Scedosporium species directly from clinical samples. The performance of the assay was then evaluated using sputum specimens from adult patients with CF.
MATERIALS AND METHODS
Fungal isolates.
To develop and validate the herein-reported species-specific primers and multiplex PCR protocol, the following fungal cultures were used: S. prolificans (strains WM 06.399 and FMR 7248), S. apiospermum (strains WM 06.472 and FMR 8619), S. aurantiacum (strains WM 06.482 and FMR 8630), S. dehoogii (strain FMR 6921), P. boydii (strain FMR 4167), P. minutispora (strain FMR 4072), Candida albicans (strain WM 06.63), Candida parapsilosis (strain WM 04.547), Candida krusei (strain WM 04.541), Cryptococcus neoformans (strain WM 08.26), Fusarium oxysporum (strain WM 08.29), Penicillium chrysogenum (strain WM 06.341), Aspergillus flavus (strain WM 06.355), Aspergillus fumigatus (strain WM 06.357), Exophiala dermatitidis (strain WM 07.305), Paecilomyces lilacinus (strain WM 04.473), Acremonium kiliense (strain WM 04.492), and Rhizopus oryzae (strain WM 10.160). All isolates were obtained from the culture collection of the Molecular Mycology Research Laboratory, Sydney Medical School—Westmead Hospital, Sydney, Australia, and subcultured onto Sabouraud dextrose agar (Oxoid, Hampshire, United Kingdom) at room temperature for 5 days to ensure adequate growth and purity prior to use.
DNA extraction.
Extraction of genomic DNA was performed as previously described (22).
Sputum sampling and extraction.
Two hundred and eight expectorated sputum samples were obtained from 69 patients attending the Westmead Hospital Adult CF Clinic, Westmead, NSW, Australia, from April 2008 to March 2009. Approval for the study was obtained from the human ethics review board of the hospital (Sydney West Area Health Service approval no. 08/WMEAD/53).
Samples were cultured on nonselective (Sabouraud dextrose agar with antibiotics [SABD]) and selective mycological (Mycosel and Scedosporium-selective [SceSel+]) media as previously described (1). Plates were incubated at 30°C and examined twice weekly for 28 days. Scedosporium spp. were identified by standard morphological and phenotypic methods (11, 13). Isolates were further characterized as S. prolificans, S. aurantiacum, or P. boydii species complex/P. apiosperma by restriction fragment length polymorphism (RFLP) analysis of the ITS region with the enzymes Sau96I and HhaI as previously described (10).
DNA was extracted from respiratory samples by using the NucliSens easyMAG automated DNA/RNA isolation platform (bioMérieux, Boxtel, Netherlands) according to the manufacturer's protocol. Extracts were stored at −20°C during the study period until used for multiplex PCR.
Primer design.
Species-specific primers were designed based on the sequences of the ITS regions (ITS1 and ITS2) and 28S ribosomal DNA gene from the members of the Pseudallescheria/Scedosporium species complex as well as other clinically important fungi, obtained from the GenBank database. The downloaded sequences were aligned to identify unique locations for the Scedosporium species-specific primer design. The specificity of the designed primers was tested using the program Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi). The species-specific primer pairs designed in this study are listed in Table 1.
Table 1.
Scedosporium species-specific primer pairs developed and evaluated as part of this study
| Species | Primer name | Primer sequence | Amplicon size (bp) | Primer pairs used in: |
|---|---|---|---|---|
| P. boydii species complex/P. apiosperma | MSAP1 | 5′ AGGCCTGCCGTCAAACCACCTAAC 3′ | 300 | Species-specific single PCR and multiplex PCR |
| MSA2 | 5′ CTACTCGACTCGTCGAAGGAGC 3′ | 300 | Species-specific single PCR and multiplex PCR | |
| P. minutispora | MPMI1 | 5′ CTCTGTTTCGTTAGCGAAAGCTCAG 3′ | 450 | Species-specific single PCR only |
| MSA2 | 5′ CTACTCGACTCGTCGAAGGAGC 3′ | 450 | Species-specific single PCR only | |
| S. aurantiacum | MSAU1 | 5′ TAGCGGATTACAAGCTCTGATTG 3′ | 650 | Species-specific single PCR and multiplex PCR |
| MSA2 | 5′ CTACTCGACTCGTCGAAGGAGC 3′ | 650 | Species-specific single PCR and multiplex PCR | |
| S. dehoogii | MSDE1 | 5′ CGCCCGAAAGGACGACGGC 3′ | 700 | Species-specific single PCR only |
| MSA2 | 5′ CTACTCGACTCGTCGAAGGAGC 3′ | 700 | Species-specific single PCR only | |
| S. prolificans | MSPF1 | 5′ CATTACCGAGTTATTACTCCAAACC 3′ | 180 | Species-specific single PCR and multiplex PCR |
| MSPR2 | 5′ TCCGTTGTTGAAAGTTTTGATTTGT 3′ | 180 | Species-specific single PCR and multiplex PCR |
Single PCR amplification.
Individual PCRs for each species-specific primer pair were performed to verify the species specificity. The PCRs were performed in a volume of 25 μl, with each reaction mixture containing 1× PCR buffer (20 mM Tris-HCl, 50 mM KCl), 1.5 mM MgCl2, 0.2 mM each deoxyribonucleotide triphosphates (dATP, dCTP, dGTP, dTTP) (Invitrogen, Carlsbad), 0.4 mM each primer (see Table 1), and 1.25 U of Taq DNA polymerase (Bioline, London, United Kingdom). Fifty nanograms of DNA or 5 μl of sputum extract was added to the reaction mixture. Sterile molecular-biology-grade water in place of DNA/sputum extract was used as a negative control, and genomic DNA extracted from the type cultures of P. apiosperma strain WM 08.194 (FMR 8619, CBS 117407), S. aurantiacum strain WM 08.202 (FMR 8630, CBS 116910), and S. prolificans strain WM 09.399 was used as a positive control in each reaction batch. The PCR amplification was performed in a thermal cycler (Perkin Elmer Cetus, Norwalk, CT) using the following conditions: an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 45 s, 65°C for 45 s, 72°C for 1 min, and a final extension step at 72°C for 10 min. The PCR products were separated on 1.4% agarose gels in Tris-borate-EDTA (TBE) buffer and visualized by UV transillumination.
Multiplex PCR.
The multiplex PCR was performed using the same protocol as was used for the individual PCRs described above, with an equimolar amount of each of the three primer sets (listed in Table 1) added to the reaction mixture. The PCR amplifications were done using the same cycling conditions as described above. The resulting amplicons were separated on 1.4% agarose gels and visualized by UV transillumination.
Assay detection limit.
DNA extract from cultured S. aurantiacum strain WM 08.202 was serially diluted in sterile water. The PCR assay was performed as described above using a DNA template concentration from 1 fg to 1 ng per PCR. The PCR products were detected as described above. The lowest DNA concentration that produced a visible band was considered the detection limit of this assay.
RESULTS
Single PCR.
Using extracted DNA from pure cultures as a template, the individual single-species-specific PCRs produced species-specific band sizes (Table 1, Fig. 1). No cross amplification of any of the species-specific primers with the other Scedosporium species was observed (data not shown). Figure 2 illustrates the results obtained by the developed multiplex PCR assay when tested on the targeted Scedosporium species and examples of a range of non-Scedosporium fungi that could be expected to be present in CF sputa. The primers did not amplify fungal DNA from the other fungal species tested (Candida albicans, Candida parapsilosis, Candida krusei, Cryptococcus neoformans, Fusarium oxysporum, Penicillium chrysogenum, Aspergillus flavus, Aspergillus fumigatus, Exophiala dermatitidis, Paecilomyces lilacinus, Acremonium kiliense, Rhizopus oryzae). A serial dilution of extracted human DNA was tested with the assay to investigate possible cross-reactivity with human genomic DNA. No cross-reactivity with human DNA was observed when tested with any of the individual or combined primer pairs (data not shown). The assay yielded 100% specificity when tested on DNA extracted from pure fungal cultures with a detection limit of 100 fg of DNA (data not shown).
Fig. 1.

Species-specific single PCR assay performed on DNA extracted from pure cultures. Lanes: 1, S. dehoogii amplified with the primer pair MSDE1/MSA2; 2, S. aurantiacum amplified with the primer pair MSAU1/MSA2; 3, P. minutispora amplified with the primer pair MPMI1/MSA2; 4, P. boydii species complex/P. apiosperma amplified with the primer pair MSAP1/MSA2; 5, S. prolificans amplified with the primer pair MSPF1/MSPR2; M, 1-kb-plus DNA marker (Invitrogen, Carlsbad, CA).
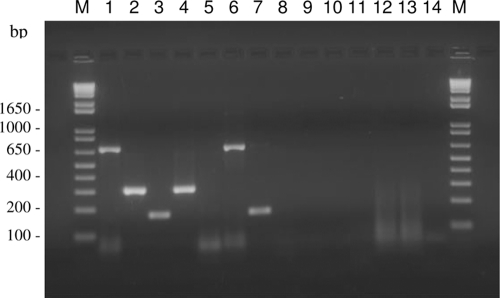
Fig. 2.
Specificity evaluation of the multiplex PCR assay performed on DNA extracted from pure Scedosporium sp. cultures (lanes: 1, Scedosporium aurantiacum strain WM 08.202 [FMR 8630]; 2, S. apiospermum strain WM 08.194 [FMR 8619, CBS 117407]; 3, S. prolificans strain WM 06.399; 4, S. apiospermum strain WM 06.472; 5, S. dehoogii strain FMR 6921; 6, S. aurantiacum strain WM 06.482; 7, S. prolificans strain FMR 7248) and examples of non-Scedosporium fungal species possibly detected in CF sputa (lanes: 8, Aspergillus fumigatus strain WM 06.357; 9, Aspergillus flavus strain WM 06.355; 10, Candida parapsilosis strain WM 04.547; 11, Candida krusei strain WM 04.541; 12 and 13, Candida albicans strain WM 06.63; 14, negative control; M, 1-kb-plus DNA marker [Invitrogen, Carlsbad, CA]).
Multiplex PCR.
In the multiplex PCR, no overlapping amplifications between the different primer pairs of S. aurantiacum, S. prolificans, and P. boydii species complex/P. apiosperma were obtained (Fig. 2). The same species-specific bands were generated as had been obtained in the individual PCRs for each species (data not shown). However, cross-amplification occurred between the S. dehoogii- and P. minutispora-specific primer pairs with the other Scedosporium species-specific primers when used in a multiplex setting, leading to the exclusion of these species from the multiplex PCR assay.
The multiplex PCR assay was validated using DNA extracted from 208 respiratory samples (sputa) from 69 patients. All positive results were repeated at least twice to exclude human or technical error. The sensitivity and specificity of the assay were calculated against the identification results obtained from culture as the gold standard. Nonselective and/or selective culture isolated Scedosporium spp. in 29 sputum samples (13.9%; S. aurantiacum, 15 samples; S prolificans, 10 samples; P. boydii species complex/P. apiosperma, 4 samples) from 11 patients. Scedosporium spp. were detected by PCR in 18 of the 29 culture-positive specimens (sensitivity, 62.1%) (Table 2) and in 7 of the 11 patients (sensitivity, 63.3%). Eleven false-negative PCRs were noted from four patients; these included six samples from one patient.
Table 2.
Test performance parameters of the PCR based on the number of specimens (and number of patients)
| Species | % |
|||
|---|---|---|---|---|
| Sensitivity | Specificity | Positive predictive value | Negative predictive value | |
| S. aurantiacum (n = 15) | 60.0 | 100 | 100 | 97.0 |
| S. prolificans (n = 10) | 70.0 | 99.0 | 77.8 | 98.5 |
| P. boydii/S. apiospermum (n = 4) | 50.0 | 98.5 | 40.0 | 99.0 |
| All Scedosporium spp. (n = 29) | 62.1 (63.6) | 97.2 (98.3) | 78.3 (87.5) | 94.1 (93.4) |
The multiplex assay had an excellent specificity for the three targeted Scedosporium species, with accurate species identification in all cases (data not shown). Multiplex PCR resulted in positive results in five culture-negative samples. The specificity of the developed multiplex PCR assay was 100% when performed on pure culture and 97.2% when performed on sputum extracts (Table 2), with a positive predictive value (PPV) of 78.3% and a negative predictive value (NPV) of 94.1% (Table 2). When the test was analyzed according to the number of patients, no significant changes in the obtained values were observed (see values given in parentheses in Table 2).
Bacterial and non-Scedosporium fungal cocolonization had no significant influence on the proportion of false-positive or false-negative samples: bacterial colonization and non-Scedosporium fungal colonization were identified in 13/18 (72.2%) and 13/18 (72.2%) of true positives, 5/5 (100%) and 2/5 (40%) of false positives, 11/11 (100%) and 11/11 (100%) of false negatives, and 30/174 (17.2%) and 141/174 (81.0%) of true negatives. Specimen processing time (after sputum storage for less than 6 months or more than 6 months) had no significant influence on the proportion of false-positive or false-negative samples (data not shown).
DISCUSSION
Scedosporium species are increasingly important fungal pathogens. Timely and accurate detection of these infective agents in clinical specimens is critical to the appropriate management of affected patients. PCR-based detection assays for pathogenic fungi have been widely used. It is important for these assays to be robust and to be easily performed in the routine mycology laboratory. The present study describes the design of five species-specific PCR primers and the subsequent development and validation of a multiplex PCR assay using three of the five species-specific primer pairs to detect and identify clinically important Pseudallescheria/Scedosporium species in respiratory (sputum) specimens. The ITS region was used as the basis for the primer design due to its presence as a multicopy gene in the fungal genome (>100 copies). Even though this region is highly conserved, it is adequately variable to enable species differentiation within the Pseudallescheria/Scedosporium species complex. The multiplex PCR assay showed a sensitivity and specificity of 100% using DNA from pure cultures, with clear and accurate differentiation of the three major pathogens—S. prolificans, P. boydii species complex/P. apiosperma, and S. aurantiacum. Other common fungal pathogens were not amplified, thus demonstrating the high specificity of this assay.
To evaluate the performance and clinical applicability of this assay, we tested its ability to detect and identify Scedosporium spp. in a large number (n = 208) of sputum specimens collected from CF patients. These patients were studied since they have a propensity to be colonized by filamentous fungi, including Scedosporium species (6, 9, 28), thus providing a suitable test population for the assay evaluation. When tested on expectorated sputum sample extracts, the multiplex PCR assay correctly identified the targeted Scedosporium species with an overall sensitivity of 63.61% in the studied patients. This result is comparable to those found for PCR assays developed for the direct detection of fungal pathogens in clinical samples. For example, an Aspergillus PCR assay showed a sensitivity ranging from 40% to 100% when performed on clinical specimens, including blood, bronchoalveolar lavage (BAL) fluid, fine-needle aspiration, or biopsy specimen (18). In a pan-fungal PCR assay including the detection of S. prolificans, the sensitivities were reported to be either 97% or 68% when applied on fresh tissue specimen or paraffin-embedded specimen, respectively (19). However, a study using an oligonucleotide array assay for the direct detection of fungi in sputum samples from a similar group of patients showed a sensitivity and specificity of 100% and 99%, respectively (3), which is most likely due to the different assay format.
Reviewing the false-negative PCRs, we noted that six of these samples, which grew S. aurantiacum on culture, were serial samples belonging to the same patient. The possible factors contributing to the persistent negative reactions in this particular patient are unknown. An unexplainable PCR negativity was also observed for culture-positive BAL specimens in a previously reported study (18). Possible explanations for these and the other five false-negative results could include insufficient amounts of Scedosporium DNA in the extracted sputa or the presence of endogenous PCR inhibitors. Inhibition of PCR in sputum samples following certain DNA isolation procedures has been reported previously for other PCR-based assays (23, 27).
The overall specificity of this assay in the patient cohort studied was 98.3% (Table 2), whereas the positive predictive value (PPV) and the negative predictive value (NPV) were 87.5% and 93.4%, respectively. False-positive results for the detection of S. apiospermum were noted in five clinical samples, of which four samples belonged to a patient who had positive cultures in other samples (not subjected to PCR amplification, due to an insufficient amount of sputum). Nevertheless, we noted earlier that the assay specificity was 100% when used on DNA extracted from pure cultured isolates. The high specificity provides a great advantage as to specifically identifying S. prolificans, P. boydii species complex/P. apiosperma, and S. aurantiacum. This is particularly important, as it will help in confirming the identities of phenotypically atypical strains, a phenomenon not uncommonly observed among the clinical isolates in this study. Species identification is further important, as species-specific clinical associations and antifungal susceptibilities have been described (12, 16). The ability to detect the three major clinically important Scedosporium species directly in sputum samples will substantially reduce the turnaround time for microbiological species identification, which will in turn allow a more timely response and better patient management.
In conclusion, this study developed species-specific primer pairs to be used either individually or in a multiplex PCR assay for the identification of the major clinically relevant Scedosporium species from pure culture and for the direct detection in sputum samples. The herein-developed PCR assay can be performed in routine clinical laboratories with a basic PCR setup. This single-step assay allowed for a high analytical sensitivity with a detection limit of as low as 100 fg Scedosporium DNA and with a high specificity. The assay can be used as a complement to current culture and morphological-based identification methods to reduce the turnaround time and to increase the detection rate. Even though the assay was evaluated on CF samples, it could equally be applied to other patient groups. Further evaluation of this assay using other clinical specimens, including blood and tissue, is indicated to determine its position as a diagnostic tool.
ACKNOWLEDGMENTS
We thank Josef Guarro from the Universitat Rovira i Virgili, Reus, Spain, for providing the Scedosporium reference strains used during the development phase of the multiplex PCR assay as well as Sue Sleiman and Ok-Cha Lee from the mycology laboratory of the Centre for Infectious Diseases and Microbiology, Westmead Hospital, Sydney, Australia, who assisted with phenotypic identification.
This work was supported by NH&MRC project grant number 352303 to W.M. and research funding by Merck, Sharp & Dohme, Australia, to W.M. and S.C.-A.C. Azian Harun was supported by the Academic Staff Higher Education Scheme scholarship from the Universiti Sains Malaysia.
This article is a publication of the ISHAM working group on Pseudallescheria/Scedosporium infections.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Blyth C. C., et al. 2010. Detection of occult Scedosporium species in respiratory tract specimens from patients with cystic fibrosis by use of selective media. J. Clin. Microbiol. 48:314–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blyth C. C., et al. 2010. Clinical associations and prevalence of Scedosporium spp. in Australian cystic fibrosis patients: identification of novel risk factors? Med. Mycol. 48:S37–S44 doi:10.3109/13693786.2010.500627 [DOI] [PubMed] [Google Scholar]
- 3. Bouchara J. P., et al. 2009. Development of an oligonucleotide assay for direct detection of fungi in sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 47:142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castelli M. V., et al. 2008. Development and validation of a quantitative PCR assay for diagnosis of scedosporiosis. J. Clin. Microbiol. 46:3412–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen S. C. A., Halliday C., Meyer W. 2002. A review of nucleic acid-based diagnostic tests for systemic mycoses with an emphasis on polymerase chain reaction-based assays. Med. Mycol. 40:333–357 [DOI] [PubMed] [Google Scholar]
- 6. Cimon B., et al. 2000. Clinical significance of Scedosporium apiospermum in patients with cystic fibrosis. Eur. J. Clin. Microbiol. 19:53–56 [DOI] [PubMed] [Google Scholar]
- 7. Cooley L., Spelman D., Thursky K., Slavin M. 2007. Infections with Scedosporium apiospermum and Scedosporium prolificans in Australia. Emerg. Infect. Dis. 13:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortez K. J., Roilides E., Quiroz-Telles F., Meletiadis J., Antachopoulos C. 2008. Infections caused by Scedosporium spp. Clin. Microbiol. Rev. 21:157–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Defontaine A., Zouhair R., Cimon B., Carrere J., Bailly E. 2002. Genotyping study of Scedosporium apiospermum isolates from patients with cystic fibrosis. J. Clin. Microbiol. 40:2108–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delhaes L., et al. 2008. Molecular typing of Australian Scedosporium isolates showing genetic variability and numerous S. aurantiacum. Emerg. Infect. Dis. 14:282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilgado F., Cano J., Gene J., Guarro J. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J. Clin. Microbiol. 43:4930–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gilgado F., Serena C., Cano J., Gené J., Guarro J. 2006. Antifungal susceptibilities of the species of the Pseudallescheria boydii complex. Antimicrob. Agents Chemother. 50:4211–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gilgado F., Cano J., Gene J., Sutton D. A., Guarro J. 2008. Molecular and phenotypic data supporting distinct species statuses for Scedosporium apiospermum and Pseudallescheria boydii and the proposed new species Scedosporium dehoogii. J. Clin. Microbiol. 46:766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilgado F., Gene J., Cano J., Guarro J. 2010. Heterothallism in Scedosporium apiospermum and description of its teleomorph Pseudallescheria apiosperma sp. nov. Med. Mycol. 48:122–128 [DOI] [PubMed] [Google Scholar]
- 15. Guignard S., et al. 2008. Multifocal Scedosporium apiospermum spondylitis in a cystic fibrosis patient. J. Cyst. Fibros. 7:89–91 [DOI] [PubMed] [Google Scholar]
- 16. Heath C. H., et al. 2009. Population-based surveillance for scedosporiosis in Australia: epidemiology, disease manifestations and emergence of Scedosporium aurantiacum infection. Clin. Microbiol. Infect. Dis. 15:680–693 [DOI] [PubMed] [Google Scholar]
- 17. Husain S., et al. 2005. Infections due to Scedosporium apiospermum and Scedosporium prolificans in transplant recipients: clinical characteristics and impact of antifungal agent therapy on outcome. Clin. Infect. Dis. 40:89–99 [DOI] [PubMed] [Google Scholar]
- 18. Lass-Florl C., et al. 2004. Diagnosing invasive aspergillosis during antifungal therapy by PCR analysis of blood samples. J. Clin. Microbiol. 42:4154–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lau A., et al. 2007. Development and clinical application of a pan-fungal PCR assay to detect and identify fungal DNA in tissue specimen. J. Clin. Microbiol. 45:380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lau A., et al. 2008. Multiplex tandem PCR: a novel platform for rapid detection and identification of fungal pathogen from blood culture specimens. J. Clin. Microbiol. 46:3021–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lau A., Sorrell T. C., Lee O., Stanley K., Halliday C. 2008. Colony multiplex-tandem PCR for rapid, accurate identification of fungal cultures. J. Clin. Microbiol. 46:4058–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer W., et al. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790–1799 [DOI] [PubMed] [Google Scholar]
- 23. Nolte F. S., et al. 1993. Direct detection of Mycobacterium tuberculosis in sputum by polymerase chain reaction and DNA hybridization. J. Clin. Microbiol. 31:1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orens J. B., et al. 2006. International guidelines for the selection of lung transplant candidates: 2006 updates—a consensus report from the pulmonary scientific council of the International Society for Heart and Lung Transplantation. J. Heart Lung Transplant. 25:745–755 [DOI] [PubMed] [Google Scholar]
- 25. Pihet M., et al. 2009. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis—a review. Med. Mycol. 47:387–397 [DOI] [PubMed] [Google Scholar]
- 26. Rodriguez-Tudela J. L., Berenguer J., Guarro J. 2009. Epidemiology and outcome of Scedosporium prolificans infection, a review of 162 cases. Med. Mycol. 47:359–370 [DOI] [PubMed] [Google Scholar]
- 27. Suffys P., et al. 2001. Inhibition of polymerase chain reaction by sputum samples from tuberculosis patients after processing using a silica-guanidiniumthiocyanate DNA isolation procedure. Mem. Inst. Oswaldo Cruz 96:1137–1139 [DOI] [PubMed] [Google Scholar]
- 28. Williamson E. C., et al. 2001. Molecular epidemiology of Scedosporium apiospermum infection determined by PCR amplification of ribosomal intergenic spacer sequences in patients with chronic lung disease. J. Clin. Microbiol. 39:47–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X., et al. 2008. Practical method for detection and identification of Candida, Aspergillus, and Scedosporium spp. by use of rolling-circle amplification. J. Clin. Microbiol. 46:2423–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]