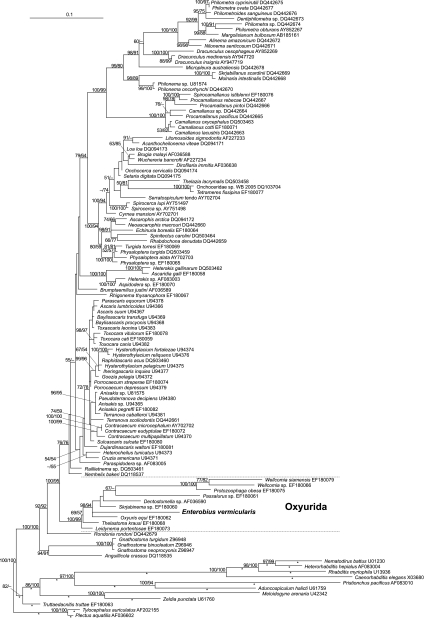
Fig. 1.
Phylogenetic placement of Enterobius vermicularis as derived from heuristic maximum likelihood analysis of an alignment of 18S rDNA sequences. The species is given first and then the GenBank (NCBI) accession number. The best tree found was rooted with Tylocephalus auriculatus and Plectus aquatilis. Branch lengths are given in terms of the estimated numbers of nucleotide substitutions per site. Asterisks mark branches that were scaled with factor 0.85 for graphical reasons. The numbers at the nodes are the maximum likelihood/maximum parsimony values. Branch support was calculated by maximum likelihood/maximum parsimony bootstrap from 1,000 replicates; values below 50% are designated by a − symbol or omitted. Bar, 0.1 nucleotide substitution per site.