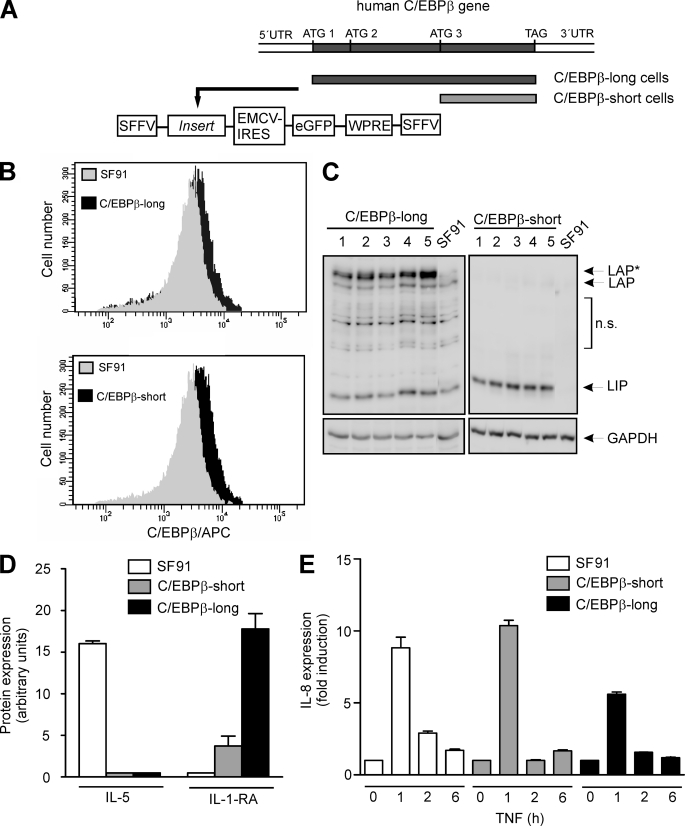
FIGURE 2.
Generation of stably transduced cells and monitoring gene expression. A, using PCR, DNA fragments were generated covering either the complete C/EBPβ coding sequence (from ATG1 to TAG; used for generation of C/EBPβ-long cells), including all three start codons for the alternative translation, or the sequence starting at the third start codon (from ATG3 to TAG), thus only coding for LIP (used for generation of C/EBPβ-short cells). The amplified fragments were cloned into the gammaretroviral vector SF91.IRES.eGFP. A schematic illustration of stably transduced functionally relevant regions containing the insert is shown. For generation of the control cell line SF91 the empty vector was used. SFFV-LTR, SFF virus long terminal repeat; EMCV-IRES, EMC virus internal ribosomal entry site; WPRE, WH virus post-transcriptional regulatory element. B, successful stable transduction of THP-1 cells was monitored using FACS analysis. Following incubation of 1 × 106 cells for 24 h, C/EBPβ proteins were tagged using the primary C/EBPβ antibody and an allophycocyanin-coupled secondary antibody. FACS analysis reveals endogenous C/EBPβ expression in SF91 control cells (gray) as well as C/EBPβ overexpression (black) in C/EBPβ-long and C/EBPβ-short cells. C, validation of C/EBPβ expression in C/EBPβ-long and C/EBPβ-short cells. Following an incubation phase for 24 h after seeding, C/EBPβ levels were measured by Western blot analysis in total cell extracts of 1 × 106 stably transduced THP-1 cells derived from five representative single cell cultures. LAP*, LAP, and LIP are indicated (loading control, GAPDH). Please note that for the blot of the extracts derived from C/EBPβ-short cells, a short exposure time was selected in order to reduce the signal of the endogenous C/EBPβ isoforms. For further experiments, the clones exhibiting the strongest C/EBPβ expression were selected (C/EBPβ-long, clone 5; C/EBPβ-short, clone 3). n.s., nonspecific bands. D, protein expression of IL-5 and IL-1-RA was determined in supernates of unstimulated SF91, C/EBPβ-short, and C/EBPβ-long cells using the proteome profiler technique (n = 4, mean ± S.D. (error bars)). E, SF91, C/EBPβ-short, and C/EBPβ-long cells were incubated with TNF up to 6 h, and the production of IL-8 mRNA was determined using real-time RT-PCR (mean ± S.D., n = 2).