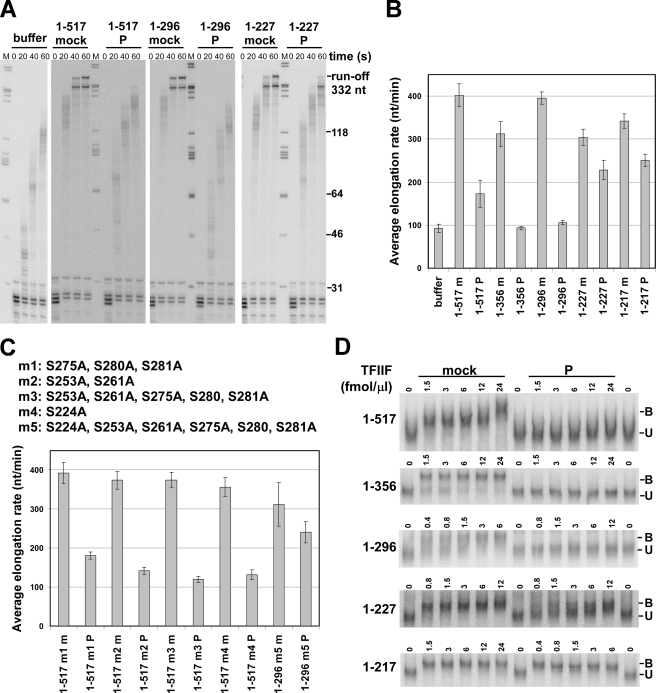
FIGURE 2.
Phosphorylation of TFIIF by CK2 impairs its activity during elongation. A, high salt-washed transcription complexes containing labeled 30-nt RNAs were chased with 0.25 mm NTPs at 30 °C for the indicated times with saturating amounts of the indicated TFIIF proteins (48 to 96 fmol/μl) or with buffer only. The positions of selected DNA molecular weight markers (M) are indicated to the right of the panel. B, average elongation rates were calculated from results like those in A by determining the average length of transcripts in lanes where the polymerase had not yet reached the end of the template. The lengths of the RAP74 subunit and the modification states of TFIIF (m, mock; P, phosphorylated) are shown. The error bars indicate mean ± S.D. based on three to nine repeats. C, RAP74 variants with the indicated Ser-to-Ala substitutions were made either with full-length RAP74 (m1-m4) or the 1–296 truncation (m5). Assays identical to those in A were performed with CK2-modified (P) or mock-phosphorylated (m) TFIIF preparations containing the indicated variants. Average elongation rates were determined as in B. The error bars indicate mean ± S.D. for three determinations. D, electrophoretic mobility shift assay with high salt-washed ternary complexes containing 30-nt labeled transcripts. Mock-treated or phosphorylated TFIIF was added to complexes at the indicated concentrations, and samples were run on 4% native acrylamide gels. The composition of the RAP74 subunit of TFIIF is indicated to the left of each gel. Unbound (U) and bound (B) complexes are labeled.