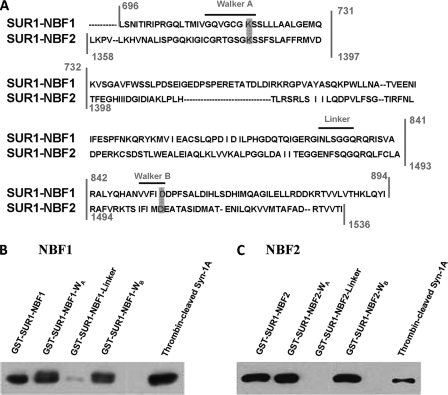
FIGURE 1.
Syntaxin-1A binds SUR1 at WA and WB regions of NBF1 and NBF2. A, SUR1-NBF1 and -NBF2 sequence alignment. The vertical lines indicate the truncation segments that we generated (see “Materials and Methods”) for each NBF, with each truncate containing WA, linker, or WB motifs. Horizontal dashes within sequences indicate a lack of residue for the respective NBF. Characteristic motifs are labeled above the sequence with horizontal lines. Notice that there is one motif in each truncated region, for a total of three truncated sections per NBF. The gray boxes indicate the specific sites we chose to mutate (see Fig. 5). B, binding of Syn-1A to SUR1-NBF1-WA and -WB domains. GST-SUR1-NBF1 (as a positive control), GST-SUR1-NBF-WA, -WB, and -linker (all bound to glutathione agarose beads, 400 pmol of protein each) were used to pull down thrombin-cleaved Syn-1A (300 μg of protein). Thrombin-cleaved Syn-1A (15 μg of protein) was used as a positive control. The precipitated proteins were separated on 15% SDS-PAGE, and the protein of interest was probed with mouse anti-Syn-1A antibody. C, binding of Syn-1A to SUR1-NBF2-WA and -WB domains. The same binding assay was performed as described in B, but WT GST-SUR1-NBF2 and truncates were used to replace WT GST-SUR1-NBF1 and truncates.