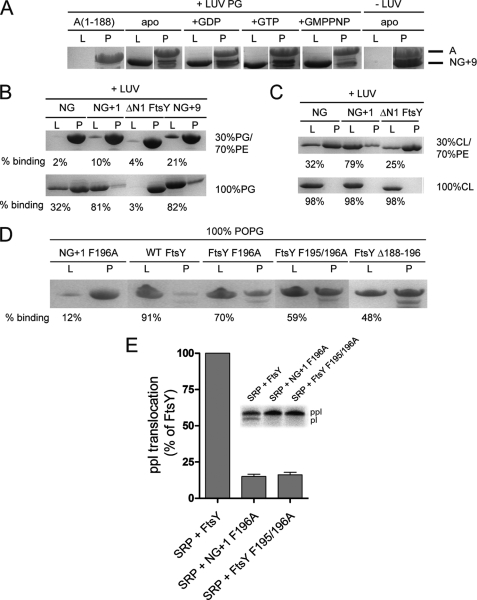
FIGURE 4.
Lipid binding properties of FtsY variants. A, the nucleotide dependence of NG+9 and A domain binding to LUVs (PG) was investigated by density gradient flotation analysis. The LUV and pellet fractions are labeled with “L” and “P,” respectively. B and C, binding of NG, NG+1, NG+9, and ΔN1 FtsY to LUVs containing different molar ratios of PG, PE, and cardiolipin (CL). The ΔN1 FtsY lacks the complete MTS sequence and was used as a negative control. D, binding of full-length FtsY and different MTS variants to LUVs (PG). The amount of protein bound to LUVs is given as percentage of total protein. For lipid binding studies, 20 μg (5–10 μm) of proteins and 1.8 mm phospholipids in the absence or presence of 2 mm nucleotides were used. E, ability of FtsY, NG+1 F196A, and FtsY F195/196A to support membrane translocation of the SRP model substrate ppl. The inset shows the SDS-PAGE used to separate ppl and prolactin (pl). The efficiency of ppl translocation was quantified and is given in percent relative to FtsY. The error bars represent the standard deviation between three independent measurements. POPG, 1-palmitoyl-2-oleoyl-phosphatidylglycerol.