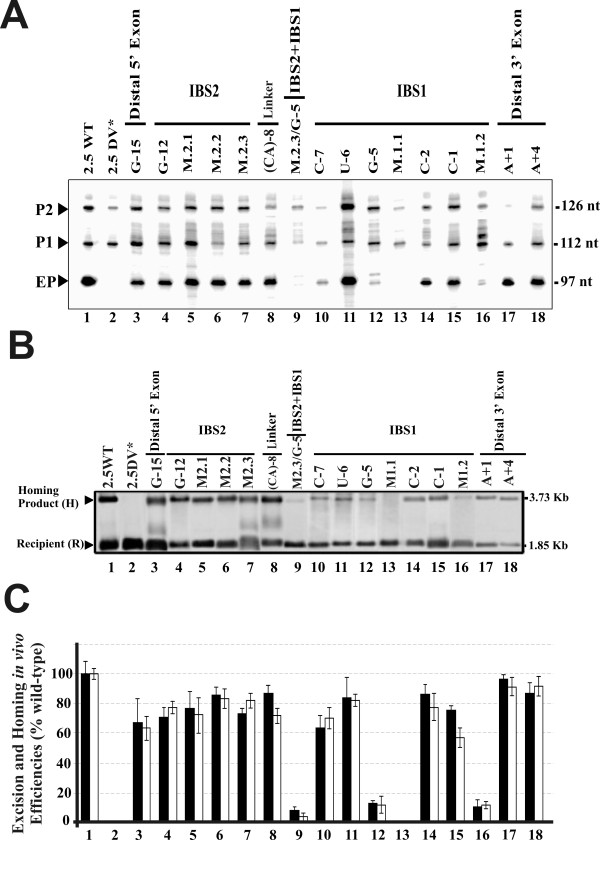
Figure 2.
Effect of exon sequence mutations in intron excision and homing efficiencies. (A) Autoradiograph of the products obtained in a primer extension assay on RNPs isolated from the S. meliloti RMO17 strain harboring the wild-type intron pKG2.5 (2.5WT), the excision-defective dV intron mutant pKG2.5D5-CGA (2.5DV*) or the exon mutations generated for this study. The RNP preparations were reverse transcribed with the intron-specific primer, P. Three main products were obtained: (EP) a 97 nt cDNA product corresponding to the excised intron RNA; and the (P1) and (P2) cDNA products (112 and 126 nt in size, respectively) that are derived from unspliced precursor transcripts, which appear to be processed at specific 5'U residues as indicated by RACE mapping in the wild-type donor construct. (B) Representative hybridization blot of homing assays. Plasmid pools from S. meliloti RMO17 harboring the wild-type pKG2.5 (2.5WT), the excision-defective dV intron mutant pKG2.5D5-CGA (2.5DV*) or the exon mutated intron donors with the pJB0.6LAG target-recipient plasmid were analyzed by SalI digestion and Southern hybridization with an exon-specific probe. The hybridization signals and the size corresponding to the target recipient plasmid (R) and the homing product (H) are indicated. Donor plasmid hybridization signal were removed. (C) Bar graph showing the excision (black bars) or homing (white bars) efficiencies, relative to the wild-type, into each of the intron constructs generated, calculated as indicated in Methods. Data are the means of determinations in at least four independent assays with the SD indicated by thin lines.