Abstract
Background
Lung cancer is a major cause of death worldwide. Gene promoter methylation is a major inactivation mechanism of tumor-related genes, some of which can be served as a biomarker for early diagnosis and prognosis evaluation of lung cancer.
Methods
We determined the promoter methylation of 6 genes using quantitative methylation-specific PCR (Q-MSP) technique in 96 clinically well-characterized non-small cell lung cancer (NSCLC).
Results
Highly frequent promoter methylation was found in NSCLC. With 100% diagnostic specificity, high sensitivity, ranging from 44.9 to 84.1%, was found for each of the 6 genes. Our data also showed that promoter methylation was closely associated with histologic type. Most of genes were more frequently methylated in squamous cell carcinomas (SCC) compared to adenocarcinomas (ADC). Moreover, promoter methylation significantly increased the risk of pleural indentation in NSCLC.
Conclusion
Our findings provided evidences that multiple genes were aberrantly methylated in lung tumorigenesis, and demonstrated the promoter methylation was closely associated with clinicopathologic characteristics of NSCLC. More importantly, we first revealed promoter methylation may be served as a potentially increased risk factor for pleural indentation of NSCLC patients.
Background
Lung cancer is a leading cause of cancer death worldwide, accounting for 30% of all cancer-related deaths [1]. The 5-year survival rate between 1996-2004 is 16%-significantly lower than that of other major cancers [2]. Owing to the rapid industrialization and increase in the smoking consumption in society, lung cancer presents as the number one cancer type of threats in China [3]. Epidemiological evidence has documented that approximately 41.8 men and 19.3 women per 100,000 Chinese individuals died of lung cancer in 2005 [4]. Lung cancer is generally classified into two major histological categories, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). The latter accounts for approximately 85% of lung cancer [2]. Approximately 25-33% of NSCLC patients present with stage I or II disease, which permits surgical resection with curative intent. However, despite surgery, approximately 30-40% of patients with NSCLC who have discrete lesions and histologically negative lymph nodes die of recurrent disease [5]. Despite the fact that the cause of most lung cancer is well know, the disease has proven difficult to diagnosis early and treat successfully, reflecting limited advances in our understanding of the molecular mechanisms underlying lung carcinogenesis and individual susceptibility to lung cancer.
In addition to genetic factors [6-8], epigenetic alterations play an important role in lung cancer development and result in changes in gene function [9]. DNA methylation is an epigenetic event whose pattern is altered frequently in a wide variety of human cancers, including promoter-specific hypermethylation as well as genome-wide hypomethylation [10,11]. Gene promoter hypermethylation is among the earliest and most common alterations in human cancers, including NSCLC, which leads to gene silencing and inactivation [12,13].
In the present study, we sought to identify DNA methylation profiles in NSCLC and their association with known or suspected cancer risk factors. For this, we used quantitative methylation-specific PCR (Q-MSP) to evaluate promoter methylation of a panel of cancer-associated genes in a large cohort of clinically well-characterized NSCLC samples, including calcitonin-related polypeptide alpha (CALCA), E-cadherin (CDH1), death-associated protein kinase 1 (DAPK1), iroquois homeobox 1 (IRX2), TIMP metallopeptidase inhibitor 3 (TIMP3), and paired box 6 (PAX6). These genes were potentially important in NSCLC, some of which have been assessed by others but some of which have not been evaluated previously.
Methods
Study subjects and DNA isolation
Ninety-six tumor samples and 15 nonmalignant lung samples were obtained from NSCLC patients who underwent curative resection at the First Affiliated Hospital of China Medical University, with approval by the institutional review board of the Hospital. None of these patients received chemotherapy and radiotherapy before the surgery. Informed consent was obtained from each patient before the surgery. All of the samples were histologically examined by a pathologist at Department of Pathology of the Hospital to identify the type and other clinical characteristics of the tumors. The clinical files of these patients are shown in Table 1. Samples were prepared and genomic DNA was isolated from paraffin-embedded samples as previously described [14]. Briefly, after a treatment for overnight at room temperature with xylene to remove pareffin, tissues were digested with 1% sodium dodecyl sulfate (SDS) and 0.5 mg/ml proteinase K at 48°C for 48 h, with addition of several spiking aliquots of concentrated proteinase K to faciliate digestion. DNA was subsequently isolated using standard phenol/chloroform protocol, and was dissolved in distilled water and stored at -80°C until use.
Table 1.
Clinicopathologic characteristics of NSCLC cases
| Characteristics | No. of patients (%) |
|---|---|
| Gender | |
| Male | 66 (69) |
| Female | 30 (31) |
| Age (mean years ± S.D.) | 58.9 ± 9.2 |
| ≤ 60 | 56 (58) |
| > 60 | 40 (42) |
| Smoking history (pack-years) | |
| 0 | 30 (31) |
| 1-39 | 35 (37) |
| ≥ 40 | 31 (32) |
| Tumor size (mean cm ± S.D.) | 3.9 ± 1.7 |
| 1-3 | 37 (38) |
| 3-5 | 44 (46) |
| > 5 | 15 (16) |
| Histologic type | |
| Adenocarcinoma (ADC) | 30 (31) |
| Brochoalveplar cell (BAC) | 6 (6) |
| Adenocarcinoma (non-BAC) | 24 (25) |
| Squamous (SCC) | 66 (69) |
| Histologic stage | |
| I | 54 (56) |
| II | 32 (34) |
| III | 10 (10) |
| Lymph node metastasis | |
| No | 70 (73) |
| Yes | 26 (27) |
| Pleural indentation | |
| No | 75 (78) |
| Yes | 21 (22) |
| Invasion or Adhesion | |
| No | 65 (68) |
| Yes | 31 (32) |
Sodium bisulfite treatment
DNA from the primary tumors and nonmalignant lung samples was subjected to bisulfite treatment as described previously [14]. Briefly, a final volume of 20 μl of H2O containing 1-2 μg genomic DNA, 10 μg salmon sperm DNA, and 0.3M NaOH was incubated at 50°C for 20 min to denature the DNA. The mixture was then incubated for 2-3 h at 70°C in 500 μl of a freshly prepared solution containing 3 M sodium bisulfite (Sigma, Saint Louis, MO), 10 mM hydroquinone (Sigma, Saint Louis, MO). Subsequently, the DNA was recovered by a Wizard DNA Clean-Up System (Promega Corp., Madison, WI) following the instructions of the manufacturer, followed by ethanol precipitation, and resuspension in 30 μl of deionized H2O. After bisulfite processing, all unmethylated cytosine residues converted to uracil, whereas the methylated cytosine residues remained unchanged. Bisulfited-modified DNA samples were stored at -80°C until use.
Quantitative methylation-specific PCR (Q-MSP) analysis
After sodium bisulfite conversion, the methylation analysis was performed by the fluorescence-based quantitative PCR assay as described previously [14]. Briefly, the Q-MSP amplification was carried out in triplicate for each samples in a final reaction mixture of 20 μl containing 3 μl bisulfite-treated DNA, 600 nM each primer, 200 nM TaqMan probe, 5.5 mM MgCl2, 1 U platinum Taq polymerase, 200 μM each of deoxyguanosine triphosphate, and 2% Rox reference. After an initial denaturation step at 95°C for 2 min, 40 cylces of 15 sec at 95°C and 60 sec at 60°C for annealing and extension were run using an ABI 7500 Fast Real-Time PCR System (Foster City, CA). Normal leukocyte DNA was methylated in vitro with Sss I methylase (New Engliand Biolabs, Beverly, MA) to generate completely methylated DNA as a positive control. Each plate contained triplicate samples and multiple water blanks, as well as serial dilutions of positive methylated control to construct the standard curve. The internal reference gene β-actin was used to normalize the amount of input DNA. The primers and TaqMan probes used in the present study were presented in Table 2. The relative degree of methylation of each sample was calculated using the method described previously [15].
Table 2.
Quantitative methylation-specific PCR primer and TaqMan probe sequences used in the present study
| Genes | Forward primer sequence (5'→3') | Probe sequence (5'→3') | Reverse primer sequence (5'→3') |
|---|---|---|---|
| CALCA | GTTTTGGAAGTATGAGGGTGACG | 6FAM-ATTCCGCCAATACACAACAACCAATAAACG-TAMRA | TTCCCGCCGCTATAAATCG |
| CDH1 | AATTTTAGGTTAGAGGGTTATCGCGT | 6FAM-CGCCCACCCGACCTCGCAT-TAMRA | TCCCCAAAACGAAACTAACGAC |
| DAPK1 | GGATAGTCGGATCGAGTTAACGTC | 6FAM-TTCGGTAATTCGTAGCGGTAGGGTTTGG-TAMRA | CCCTCCCAAACGCCGA |
| IRX2 | GCGGGTCGTTTAGGTTAGTATTCG | 6FAM-CCCTCCATCCACGCCCGACCGAAA-TAMRA | CGCCGAACAACGAACAAAACG |
| TIMP3 | GCGTCGGAGGTTAAGGTTGTT | 6FAM-AACTCGCTCGCCCGCCGAA-TAMRA | CTCTCCAAAATTACCGTACGCG |
| PAX6 | ATATAGGACGGCGGTTTAGGTTG | 6FAM-CCCAAAATCCGACCGACTCCAACCCCTA-TAMRA | TTCCGACCGAACGAAAACCTAC |
| β-Actin | TGGTGATGGAGGAGGTTTAGTAAGT | 6FAM-ACCACCACCCAACACACAATAACAAACACA-TAMRA | AACCAATAAAACCTACTCCTCCCTTAA |
Statistical analysis
Promoter methylation was considered present if the ratio was above a certain cut-off value. The relative level of methylation varied significantly among the 6 genes and therefore cut-off points were studied for each gene individually. To set up cut-off value for each gene for detection of NSCLC, we used the Medcalc Software (MedCalc Software bvba, Belgium) to construct receiver operating characteristic (ROC) curves. The area under the curve of ROC curve is a measure of the ability of a continuous marker to accurately classify tumor and non-tumor tissue. Such a curve is a plot of sensitivity vs. 1 minus specificity values associated with all dichotomous markers that can be formed by varying the value threshold (or cut-off value) used to designate a marker "positive". Factors associated with patients and tumor characteristics were assessed univariately with chi-square tests for trend and logistic regression. In the final analysis, multivariable adjustments were made to adjust for the potentially confounding effects of smoking history, histologic type, lymph node metastasis, and pleural indentation. P < 0.05 was considered to be statistically significant. All statistical analyses were performed using the SPSS statistical package (11.5, Chicago, IL, USA).
Results
Demographics
In the present study, we chose genes which were potentially important in NSCLC and determined the promoter methylation of these 6 genes using Q-MSP assay in 96 well-characterized NSCLC patients. As shown in Table 1 the mean age of the 96 NSCLC cases was 58.9 years. Males were more than females (69% vs. 31%). Sixty-nine percent of patients reported a history of smoking, with 32% reporting at least 40 life-time pack-years of smoking. Ninety percent of NSCLC cases had surgical stage I or II disease and 84% had tumors < 5 cm. By histology, 31% of tumors were adenocarcinomas (ADC), including 6% bronchioloalveolar carcinomas (BAC) and 25% non-BAC adenocarcinomas, and 69% were squamous cell carcinomas (SCC). The cases with lymph node metastasis, pleural indentation and invasion or adhesion were in 26/96 (27%), 21/96 (22%), and 31/96 (32%), respectively.
Frequent promoter methylation in NSCLC
We examined promoter methylation of CALCA, CDH1, DAPK1, IRX2, TIMP3, and PAX6 using Q-MSP in a cohort of 96 NSCLC. As shown in Figure 1, the overall methylation level of each gene was higher in tumor tissues than in nonmalignant lung tissues. Among the 6 genes examined, statistical significances were observed in CALCA (P < 0.01), CDH1 (P < 0.01), and PAX6 (P < 0.001). We set up appropriate cut-off values to distinguish NSCLC from nonmalignant lung tissues, and determine diagnostic sensitivity and specificity. As shown in Figure 2, with 100% diagnostic specificity for each of the 6 genes, the sensitivity of CALCA, CDH1, DAPK1, IRX2, TIMP3, and PAX6 was 69.2%, 45.8%, 84.1%, 52.3%, 44.9%, and 75.7%, respectively.
Figure 1.
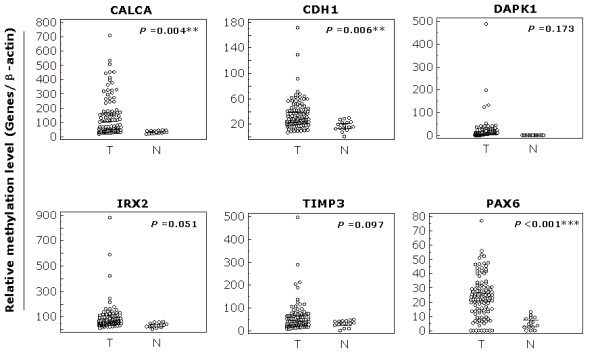
Distribution patterns of promoter methylation of the 6 genes in NSCLC. Q-MSP was performed as described in Materials and Methods. The relative methylation level (on Y axis) is represented by ratios of candidate gene/β-actin × 1000, except for PAX6/β-actin × 100. Horizonal lines indicate a 95% normal confidence interval for the sample mean. T, tumor tissues; N, nonmalignant lung tissues. **, P < 0.01; ***, P < 0.001.
Figure 2.
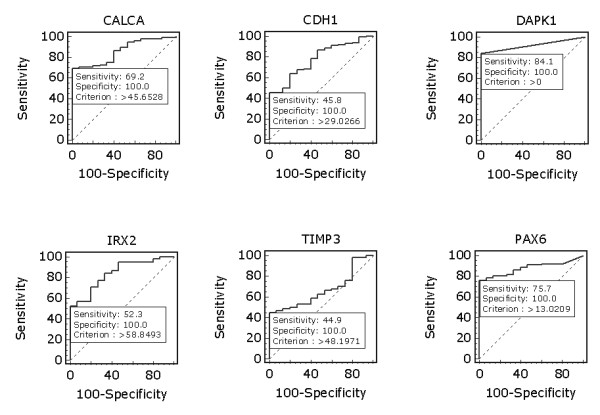
Receiver operating characteristic (ROC) curves for the 6 genes in NSCLC. All tumor and nonmalignant lung tissues for which there was complete DNA methylation data were used for the analyses. The ROC curves plot sensitivity vs. 100-specificity. The determined cut-off values for CALCA, CDH1, DAPK1, IRX2, TIMP3, and PAX6 were 45.7, 29.0, 0, 58.8, 48.2, and 13.0, respectively.
Association of promoter methylaton with clinicopathological charactristics of NSCLC
Among all clinicopathologic characteristics, methylation levels varied substantially by histologic type (Table 3). Methylation level of most of genes was higher in SCC than in ADC, particularly in CDH1 gene (P < 0.05) (Table 3). There was a significant difference in the methylation level of DAPK1 gene between lymph node metastasis group and nonmetastasis group (40.3 ± 100.5 in the former vs. 12.3 ± 18.5 in the latter, P < 0.05) (Table 3). The methylation level of CALCA gene was significant higher in the patients with invasion or adhesion compared with patients without invasion or adhesion (174.5 ± 160 vs. 115.1 ± 122.5, P < 0.05) (Table 3). The methylation level of CALCA gene was found to be significantly associated with smoking history (155.2 ± 150.2 in smokers vs. 89.6 ± 92.7 in non-smokers, P < 0.05) (Table 3). Our data showed that the methylation level of most of genes was not associated with quantity of cigarette smoking (Pack-years) (Additional file 1). Of note, there was a significant difference in the methylation level of PAX6 gene between patients with age > 60 years and ≤ 60 years (28.6 ± 16.2 in the former vs. 20.2 ± 12.4 in the latter, P < 0.05). In addition, the methylation level of CALCA gene was also found to be significantly associated with tumor stage (217.7 ± 186.7 in patients with histoligical satge ≥ III vs. 125 ± 129 in patients with histological stage < III, P < 0.05) (Additional file 1). The methylation level of several genes was associated with increased tumor size, including CALCA, DAPK1, IRX2, and TIMP3 genes, although these associations did not reach statistical difference (Additional file 1).
Table 3.
Association of the level of promoter methylation with clinicopathologic charactristics of NSCLC
|
Genes |
Gender (n = 96) | Histologic type (n = 96) | Smoking history (n = 96) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Female | Male | P | SCC | ADC | P | Y | N | P | |
| (n = 30) | (n = 66) | (n = 66) | (n = 30) | (n = 66) | (n = 30) | ||||
| CALCA | 145.0 ± 149.6 | 112.1 ± 106.5 | 0.28 | 152.1 ± 152.1 | 96.4 ± 90.6 | 0.07 | 155.2 ± 150.2 | 89.6 ± 92.7 | 0.03* |
| CDH1 | 33.1 ± 18.7 | 31.5 ± 21.2 | 0.71 | 35.4 ± 19.2 | 26.3 ± 18.6 | 0.03* | 32.2 ± 18.7 | 33.4 ± 21.3 | 0.77 |
| DAPK1 | 18.5 ± 59.6 | 23 ± 45.2 | 0.71 | 26.2 ± 65.8 | 5.9 ± 7.6 | 0.09 | 17.4 ± 59.6 | 25.4 ± 45.2 | 0.52 |
| IRX2 | 96.4 ± 131.5 | 63.9 ± 34.8 | 0.19 | 92.3 ± 115.7 | 72.9 ± 103.6 | 0.43 | 95.3 ± 132.1 | 66.3 ± 36.1 | 0.24 |
| TIMP3 | 60.5 ± 72.8 | 50.9 ± 39.1 | 0.50 | 64.9 ± 71.7 | 41.3 ± 39.8 | 0.09 | 61.5 ± 72.8 | 48.8 ± 38.9 | 0.37 |
| PAX6 | 23.4 ± 15.1 | 24.2 ± 13.8 | 0.78 | 24.0 ± 15.5 | 23.1 ± 12.8 | 0.79 | 23.9 ± 15.6 | 23.3 ± 12.6 | 0.85 |
| Genes | Lymph node metastasis (n = 96) | Pleural indentation (n = 96) | Invasion or Adhesion (n = 96) | ||||||
| Y | N | P | Y | N | P | Y | N | P | |
| (n = 26) | (n = 70) | (n = 21) | (n = 75) | (n = 31) | (n = 65) | ||||
| CALCA | 175.7 ± 175.1 | 119.5 ± 119.1 | 0.08 | 129.9 ± 132.5 | 136 ± 140.2 | 0.86 | 174.5 ± 160 | 115.1 ± 122.5 | 0.048* |
| CDH1 | 31.0 ± 13.9 | 33.1 ± 21.2 | 0.64 | 35.3 ± 21.2 | 31.8 ± 18.9 | 0.47 | 30.7 ± 16.6 | 33.5 ± 20.7 | 0.52 |
| DAPK1 | 40.3 ± 100.5 | 12.3 ± 18.5 | 0.03* | 12.4 ± 11.9 | 21.9 ± 62.3 | 0.49 | 10.9 ± 10.6 | 24.2 ± 66.6 | 0.27 |
| IRX2 | 42.7 ± 51 | 91.2 ± 127.2 | 0.47 | 73.1 ± 38.4 | 89.9 ± 124.9 | 0.54 | 71.8 ± 48.3 | 93.1 ± 131.6 | 0.38 |
| TIMP3 | 69.3 ± 91 | 53.2 ± 51 | 0.28 | 53.9 ± 42.9 | 58.6 ± 69.2 | 0.77 | 52.7 ± 53.1 | 59.8 ± 69.1 | 0.61 |
| PAX6 | 23.2 ± 14.2 | 23.9 ± 14.9 | 0.83 | 25.2 ± 13.9 | 23.3 ± 14.9 | 0.59 | 25.8 ± 15.7 | 22.7 ± 14.1 | 0.32 |
In univariate analyses, the promoter methylation of most of genes was more frequent in SCC compared to ADC, particularly in CDH1(OR = 2.63, 95% CI = 1.05-6.60), DAPK1 (OR = 6.64, 95% CI = 1.85-23.8), IRX2 (OR = 2.89, 95% CI = 1.17-7.13), and TIMP3 (OR = 2.51, 95% CI = 1.01-6.21) (Table 4). In addition, methylation of CALCA (OR = 2.13, 95% CI = 1.01-4.47) and TIMP3 (OR = 1.88, 95% CI = 1.01-3.49) was significantly associated with histologic stage (Table 4). However, there was not significant association between promoter methylation and other clinocopathologic characteristics, including gender, age, smoking history, quantity of cigarette smoking, tumor size, lymph node metastasis, pleural indentation, and invasion or adhesion (Table 4). In order to assess the independent associations between promoter methylation and smoking history, histologic type, lymph node metastasis, and pleural indentation, we conducted multiple multivariable logistic regressions (Table 5). In multivariable analyses adjusting for potential false discovery rate associated with multiple comparisons (6 different models), methylation of most of genes ramained associated with histologic type. The promoter methylation of CDH1(OR = 6.77, 95% CI = 1.71-26.8), DAPK1 (OR = 10.7, 95% CI = 2.16-52.6), IRX2 (OR = 6.38, 95% CI = 1.17-23.0), and TIMP3 (OR = 3.37, 95% CI = 1.03-11.0) genes was significant more likely in SCC compared to ADC (Table 5). However, after adjustment for smoking history, histologic type, and lymph node metastasis, the promoter methylaton of each gene was more frequent in patients with pleural indentation compared with without pleural indentation (Table 5), particularly in CDH1(OR = 5.27, 95% CI = 1.27-21.9), DAPK1 (OR = 6.98, 95% CI = 1.06-45.8), and IRX2 (OR = 3.89, 95% CI = 1.01-15.0) genes, suggesting that promoter methylation of these genes may be a potential risk of pleural indentation in NSCLC.
Table 4.
Promoter methylation in NSCLC-univariate associations with clinicopathologic characteristics (OR† and 95% CI)
| Genes | Male vs. Female | Age (per 10 years) | Smoking history | Pack-years1 | SCC vs. ADC |
|---|---|---|---|---|---|
| CALCA | 0.73 (0.28-1.90) | 0.94 (0.58-1.52) | 1.43 (0.57-3.57) | 0.88 (0.51-1.51) | 2.21 (0.89-5.47) |
| CDH1 | 2.13 (0.86-5.22) | 0.83 (0.53-1.30) | 1.41 (0.59-3.39) | 0.91 (0.55-1.50) | 2.63 (1.05-6.60)* |
| DAPK1 | 1.45 (0.43-4.87) | 1.16 (0.60-2.24) | 2.11 (0.64-6.92) | 1.83 (0.84-3.98) | 6.64 (1.85-23.8)* |
| IRX2 | 1.29 (0.54-3.06) | 0.76 (0.48-1.19) | 1.06 (0.45-2.52) | 0.91 (0.55-1.50) | 2.89 (1.17-7.13)* |
| TIMP3 | 1.09 (0.46-2.60) | 1.09 (0.70-1.71) | 1.09 (0.46-2.60) | 1.27 (0.76-2.11) | 2.51 (1.01-6.21)* |
| PAX6 | 1.04 (0.37-2.88) | 1.19 (0.70-2.03) | 0.78 (0.27-2.25) | 0.93 (0.51-1.69) | 1.04 (0.37-2.88) |
| Genes | Tumor size2 | Lymph node metastasis | Pleural indentation | Invasion or Adhesion | Histologic stage3 |
| CALCA | 1.01 (0.55-1.88) | 1.74 (0.62-4.91) | 1.18 (0.41-3.41) | 2.44 (0.88-6.79) | 2.13 (1.01-4.47)* |
| CDH1 | 1.20 (0.67-2.13) | 1.02 (0.41-2.51) | 1.79 (0.67-4.76) | 0.96 (0.41-2.27) | 1.11 (0.62-2.02) |
| DAPK1 | 1.81 (0.72-4.54) | 5.17 (0.64-41.95) | 1.63 (0.33-8.02) | 1.70 (0.43-6.67) | 1.24 (0.50-3.08) |
| IRX2 | 1.57 (0.88-2.83) | 1.17 (0.47-2.88) | 1.37 (0.52-3.63) | 0.86 (0.36-2.01) | 1.14 (0.63-2.07) |
| TIMP3 | 1.67 (0.92-3.01) | 2.05 (0.82-5.10) | 1.16 (0.44-3.05) | 0.69 (0.29-1.66) | 1.88 (1.01-3.49)* |
| PAX6 | 0.78 (0.40-1.54) | 0.74 (0.26-2.09) | 2.04 (0.54-7.69) | 1.84 (0.61-5.56) | 1.76 (0.79-3.93) |
† OR: odds ratio with 95% confidence interval.
1 Pack-years (0; > 0 and < 40; ≥ 40).
2 Tumor size (> 1 and ≤3; > 3 and ≤5; > 5).
3 Histologic stage (I; II; III).
* Significant at P < 0.05.
Table 5.
Promoter methylation in NSCLC-multivariable models assessing smoking history, histologic type, lymph node metastasis, and pleural indentation (OR† and 95% CI)
| Genes | Smoking history | SCC vs. ADC | Lymph node metastasis | Pleural indentation |
|---|---|---|---|---|
| CALCA | 1.07 (0.40-2.88) | 2.66 (0.85-8.32) | 1.43 (0.46-4.41) | 2.19 (0.61-7.79) |
| CDH1 | 0.98 (0.37-2.58) | 6.77 (1.71-26.8)* | 0.78 (0.29-2.13) | 5.27 (1.27-21.9)* |
| DAPK1 | 0.83 (0.19-3.73) | 10.7 (2.16-52.6)* | 3.07 (0.32-29.4) | 6.98 (1.06-45.8)* |
| IRX2 | 0.69 (0.26-1.83) | 6.38 (1.77-23.0)* | 0.85 (0.31-2.33) | 3.89 (1.01-15.0)* |
| TIMP3 | 0.78 (0.30-2.02) | 3.37 (1.03-11.0)* | 1.75 (0.65-4.75) | 2.66 (0.77-9.16) |
| PAX6 | 0.67 (0.21-2.08) | 1.82 (0.50-6.57) | 0.77 (0.25-2.38) | 2.66 (0.58-12.3) |
† OR: odds ratio with 95% confidence interval, adjusted for multiple comparisions.
* Significant at P < 0.05.
Discussion
We assessed the promoter methylation of the 6 genes in a large cohort of well-characterized NSCLC subjects using Q-MSP technique in the present study, including CALCA, CDH1, DAPK1, IRX2, TIMP3, and PAX6. CALCA is known to encode a peptide hormone that plays a role in maintenance of calcium levels in blood serum and T-and B-cell regulation in certain malignancies, which is frequently methylated in multiple types of cancer [16,17]. CDH1 is a classical cadherin from the cadherin superfamily, which encodes a calcium dependent cell-cell adhesion protein. Epigenetic inactivation of CDH1 is thought to contribute to progression in cancer by increasing proliferation, invasion, and/or metastasis [18]. DAPK1 encodes a structurally unique calcium/calmodulin-dependent serine/threonine kinase which acts as a positive regulator of apoptosis. It is frequently methylated in human cancers as a tumor suppressor gene [19]. IRX2 is a member of the Iroquois homeobox transcription factor family, which is involved in developmental pattern formation in multiple organs such as the brain and heart [20]. It is highly specific for tumor-associated methylation, and little or no methylation is found in nonmalignant lung tissue [21]. TIMP3 is an angiogenesis inhibitor, and its epigenetic inactivation is associated with neovascularization and invasion in human malignancy [22]. PAX6, a transcription factor, has currently been suggested to function as a tumor suppressor in glioblastoma and to act as an early differentitation marker for neuroendocrine cells [23], which is frequently methylated in human cancers [21,24]. Similarly, our findings also showed that 3 of 6 genes had significantly higher methylation level in tumor tissues than nonmalignant lung tissues, including CALCA, CDH1, and PAX6 genes. Importantly, with 100% diagnostic specificity, excellent sensitivity, ranging from 44.9 to 84.1%, was found for each of the 6 genes. The high specificity and frequency of these methylation markers make them excellent candidates for future applications developed for early diagnosis and prognosis evaluation of lung cancer.
Given smoking plays the central role in lung cancer development, it is somewhat surprising that we did not find significant association between promoter methylation of most genes and smoking history, in agreement with most studies [25-28]. However, several previous studies have reported aberrant DNA methylation of tumor-related genes was associated with tobacco smoking [29-32]. It is possible that lung cancer as a result of tobacco smoking is a complex disease with many unique genetic and epigenetic features. Better understanding of the molecular mechanisms underlying this disease would undoubtedly improve the outcomes of patients with smoking-associated lung cancer.
Our findings of substantial differences in promoter methylation depending on histologic typing of NSCLC have been reported to some degree in the literatures. Unlike what observed in the present study, a number of others have noted that adenomatous polyposis coli (APC), cyclin D2 (CCND2), potassium voltage-gated channel, subfamily H (eag-related), member 5 (KCNH5), and runt-related transcription factor 2 (RUNX2) genes were significantly more frequently in ADC compared to SCC [33-36]. Similar to the present study, although no statistical significance was observed, promoter methylation of DAPK1 gene was detected with higher frequency in SCC compared to ADC [36]. Conversely, there was not significant difference in promoter methylation of CDH1 gene between SCC and ADC in this literature [36]. Anyway, we believe that our findings that promoter methylation of several genes is more frequent in SCC compared to ADC, particularly IRX2 gene, are new to the literature. Moreover, methylation level of DAPK1 and CALCA genes was significantly associated with lymph node metastasis and invasion/adhesion, suggesting that methylation degree of these genes might contribute to oncologic outcomes of NSCLC patients.
A very recent study showed that a number of important differences in frequency of promoter methylation in females compared to males, suggesting that promoter methylation is associated with gender [36]. However, these substantial differences have not been consistently noted in the previous literatures [35,37-39]. Similarly, promoter methylation was also not significantly associated with gender in the present study. It is possible that the differences in the promoter methylation associated with gender may be related to the geographical or cultural differences in carcinogen exposures, including cigarette smoking, dietary factors, occupational and environmental chemical exposure [40-42]. The discrepant results might also have been due to genetic differences of the study populations. A previous study indicated that patients harboring functional polymorphic variants of glutathione S-transferase pi 1 (GSTP1) had a higher risk of promoter hypermethylation of O-6-methylguanine-DNA methyltransferase (MGMT) gene [43], suggesting that functional variants in the genes involved in the folate metabolism, DNA methylation, carcinogen metabolosim and the repair of methylation may play an important role in the susceptibility to promoter methylation. However, it is possible that these differences are attributable to chance as a result of the relatively small number of the study subjects examined. Therefore, further study with more subjects will be needed.
Interestingly, multivariable analyses revealed that the promoter methylation of a number of genes had signficant higher frequency in patients with pleural indentation compared with without pleural indentation, suggesting that promoter methylation may be a potentially increased risk for pleural indentation of NSCLC patients. Pleural indentation is a well-known imaging sign on chest computed tomography (CT) that suggests a possible pleural invasion by peripheral NSCLC, particularly ADC [44,45]. A previous study showed that pleural involvement was significantly correlated with a poor prognosis in NSCLC, suggesting that pleural involvement may be one of most important factors to affect on the prognosis of NSCLC [46]. Although the degree of pleural invasion is clinically important, the accurate preoperative evaluation is sometimes difficult. CT and magnetic resonance imaging (MRI) are usually used in the evaluation of tumor extent, and diagnosis of chest wall invasion, pleural dissemination and pleural effusion can be easily made preooperatively by this way [47]. However, pleural dissemination and/or pleural effusion, which were not documented preoperatively, were sometimes revealed during operation. In addition, the accruate visceral pleural invasion can not be made with preoperative CT or MRI. In the present study, our findings suggested that promoter methylation of certain genes can increase the risk of pleural indentation in NSCLC. DNA methylation is the earliest and most frequent molecular events in human tumorigenesis. Detection of aberrant methylation using some high-senstive approaches can be thus used to predict and evaluate the pleural invovlement in NSCLC.
Conclusion
In summary, in the present study, we found a panel of methylated genes that differentiate tumor tissues from nonmalignant lung tissues, which was strongly associated with clinicopathologic characteristics of NSCLC. Importantly, our data first revealed that promoter methylation may be a potentially increased risk factor for pleural indentation of NSCLC patients.
Authors' contributions
MJ and PH conceived and designed the experiments. MJ and YZ performed the experiments. MJ and PH collected the samples and analyzed the data. BS and PH contributed reagents/materials/analysis tools. PH Wrote the paper. All authors are in agreement with the content of the manuscript and this submission.
Competing interests
The authors declare that they have no competing interests.
Supplementary Material
Table S1. Association of the level of promoter methylation with age, pack-years, tumor size and histoligic stage in NSCLC
Contributor Information
Meiju Ji, Email: jimeiju@yahoo.com.
Yong Zhang, Email: hou1105@gmail.com.
Bingyin Shi, Email: shibingy@126.com.
Peng Hou, Email: phou@mail.xjtu.edu.cn.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 30901459 and 30973372) and the Fundamental Research Funds for the Central Universities.
References
- Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Song F, He M, Li H, Qian B, Wei Q, Zhang W, Chen K, Hao X. A cancer incidence survey in Tianjin: the third largest city in China-between 1981 and 2000. Cancer Causes Control. 2008;19:443–50. doi: 10.1007/s10552-007-9105-6. [DOI] [PubMed] [Google Scholar]
- Yang L, Parkin DM, Li LD, Chen YD, Bray F. Estimation and projection of the national profile of cancer mortality in China: 1991-2005. Br J Cancer. 2004;90:2157–66. doi: 10.1038/sj.bjc.6601813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–85. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- Schwartz AG, Prysak GM, Bock CH, Cote ML. The molecular epidemiology of lung cancer. Carcinogenesis. 2007;28:507–18. doi: 10.1093/carcin/bgl253. [DOI] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsäter A, Flex A, Aben KK, de Vegt F. et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- Hou P, Ji M, Xing M. Association of PTEN gene methylation with genetic alterations in the phosphatidylinositol 3-kinase/AKT signaling pathway in thyroid tumors. Cancer. 2008;113:2440–7. doi: 10.1002/cncr.23869. [DOI] [PubMed] [Google Scholar]
- Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, Holt EH, Kiseljak-Vassiliades K, Rhoden KJ, Tolaney S, Condouris S, Tallini G, Westra WH, Umbricht CB, Zeiger MA, Califano JA, Vasko V, Xing M. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer. 2006;119:2322–9. doi: 10.1002/ijc.22110. [DOI] [PubMed] [Google Scholar]
- Brait M, Begum S, Carvalho AL, Dasgupta S, Vettore AL, Czerniak B, Caballero OL, Westra WH, Sidransky D, Hoque MO. Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2786–94. doi: 10.1158/1055-9965.EPI-08-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixão VA, Vidal DO, Caballero OL, Vettore AL, Tone LG, Ribeiro KB, Lopes LF. Hypermethylation of CpG island in the promoter region of CALCA in acute lymphoblastic leukemia with central nervous system (CNS) infiltration correlates with poorer prognosis. Leuk Res. 2006;30:891–4. doi: 10.1016/j.leukres.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed IA, Pusch CM, Hamed T, Rashad H, Idris A, El-Fadle AA, Blin N. Epigenetic alterations by methylation of RASSF1A and DAPK1 promoter sequences in mammary carcinoma detected in extracellular tumor DNA. Cancer Genet Cytogenet. 2010;199:96–100. doi: 10.1016/j.cancergencyto.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF. Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol. 2000;224:263–74. doi: 10.1006/dbio.2000.9801. [DOI] [PubMed] [Google Scholar]
- Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA. 2008;105:252–7. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- Hellwinkel OJ, Kedia M, Isbarn H, Budäus L, Friedrich MG. Methylation of the TPEF-and PAX6-promoters is increased in early bladder cancer and in normal mucosa adjacent to pTa tumours. BJU Int. 2008;101:753–7. doi: 10.1111/j.1464-410X.2007.07322.x. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–6. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersting M, Friedl C, Kraus A, Behn M, Pankow W, Schuermann M. Differential frequencies of p16(INK4a) promoter hypermethylation, p53 mutation, and K-ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J Clin Oncol. 2000;18:3221–9. doi: 10.1200/JCO.2000.18.18.3221. [DOI] [PubMed] [Google Scholar]
- Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61:3419–24. [PubMed] [Google Scholar]
- Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, Shimizu N, Shimizu K, Lee H, Chen CY, Fong KM, Gilcrease M, Roth JA, Minna JD, Gazdar AF. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–60. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- Kim DH, Kim JS, Ji YI, Shim YM, Kim H, Han J, Park J. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res. 2003;63:3743–6. [PubMed] [Google Scholar]
- Toyooka S, Suzuki M, Tsuda T, Toyooka KO, Maruyama R, Tsukuda K, Fukuyama Y, Iizasa T, Fujisawa T, Shimizu N, Minna JD, Gazdar AF. Dose effect of smoking on aberrant methylation in non-small cell lung cancers. Int J Cancer. 2004;110:462–4. doi: 10.1002/ijc.20125. [DOI] [PubMed] [Google Scholar]
- Kim JS, Kim H, Shim YM, Han J, Park J, Kim DH. Aberrant methylation of the FHIT gene in chronic smokers with early stage squamous cell carcinoma of the lung. Carcinogenesis. 2004;25:2165–71. doi: 10.1093/carcin/bgh217. [DOI] [PubMed] [Google Scholar]
- de Fraipont F, Moro-Sibilot D, Michelland S, Brambilla E, Brambilla C, Favrot MC. Promoter methylation of genes in bronchial lavages: a marker for early diagnosis of primary and relapsing non-small cell lung cancer. Lung Cancer. 2005;50:199–209. doi: 10.1016/j.lungcan.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, Sidransky D. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370–5. [PubMed] [Google Scholar]
- Gu J, Berman D, Lu C, Wistuba II, Roth JA, Frazier M, Spitz MR, Wu X. Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006;12:7329–38. doi: 10.1158/1078-0432.CCR-06-0894. [DOI] [PubMed] [Google Scholar]
- Yanagawa N, Tamura G, Oizumi H, Kanauchi N, Endoh M, Sadahiro M, Motoyama T. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58:131–8. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Hawes SE, Stern JE, Feng Q, Wiens LW, Rasey JS, Lu H, Kiviat NB, Vesselle H. DNA hypermethylation of tumors from non-small cell lung cancer (NSCLC) patients is associated with gender and histologic type. Lung Cancer. 2010;69:172–9. doi: 10.1016/j.lungcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata S, Sugio K, Uramoto H, Oyama T, Hanagiri T, Morita M, Yasumoto K. The methylation status and protein expression of CDH1, p16(INK4A), and fragile histidine triad in nonsmall cell lung carcinoma: epigenetic silencing, clinical features, and prognostic significance. Cancer. 2006;106:2190–9. doi: 10.1002/cncr.21870. [DOI] [PubMed] [Google Scholar]
- Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007;110:2785–92. doi: 10.1002/cncr.23113. [DOI] [PubMed] [Google Scholar]
- Vaissiere T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, Brennan P, Tost J, Boffetta P, Herceg Z. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69:243–52. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer Res. 1996;56:3655–8. [PubMed] [Google Scholar]
- Moore LE, Huang WY, Chung J, Hayes RB. Epidemiologic considerations to assess altered DNA methylation from environmental exposures in cancer. Ann N Y Acad Sci. 2003;983:181–96. doi: 10.1111/j.1749-6632.2003.tb05973.x. [DOI] [PubMed] [Google Scholar]
- Vuillemenot BR, Pulling LC, Palmisano WA, Hutt JA, Belinsky SA. Carcinogen exposure differentially modulates RAR-beta promoter hypermethylation, an early and frequent event in mouse lung carcinogenesis. Carcinogenesis. 2004;25:623–9. doi: 10.1093/carcin/bgh038. [DOI] [PubMed] [Google Scholar]
- Gilliland FD, Harms HJ, Crowell RE, Li YF, Willink R, Belinsky SA. Glutathione S-transferase P1 and NADPH quinone oxidoreductase polymorphisms are associated with aberrant promoter methylation of P16(INK4a) and O(6)-methylguanine-DNA methyltransferase in sputum. Cancer Res. 2002;62:2248–52. [PubMed] [Google Scholar]
- Shapiro R, Wilson GL, Yesner R, Shuman H. A useful roentgen sign in the diagnosis of localized bronchioloalveolar carcinoma. Am J Roentgenol Radium Ther Nucl Med. 1972;114:516–24. doi: 10.2214/ajr.114.3.516. [DOI] [PubMed] [Google Scholar]
- Kuhlman JE, Fishman EK, Kuhajda FP, Meziane MM, Khouri NF, Zerhouni EA, Siegelman SS. Solitary bronchioloalveolar carcinoma: CT criteria. Radiology. 1988;167:379–82. doi: 10.1148/radiology.167.2.2833764. [DOI] [PubMed] [Google Scholar]
- Li M, Ito H, Wada H, Tanaka F. Pit-fall sign on computed tomography predicts pleural involvement and poor prognosis in non-small cell lung cancer. Lung Cancer. 2004;46:349–55. doi: 10.1016/j.lungcan.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Murayama S, Murakami J, Yoshimitsu K, Torii Y, Ishida T, Masuda K. CT diagnosis of pleural dissemination without pleural effusion in primary lung cancer. Radiat Med. 1996;14:117–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Association of the level of promoter methylation with age, pack-years, tumor size and histoligic stage in NSCLC


