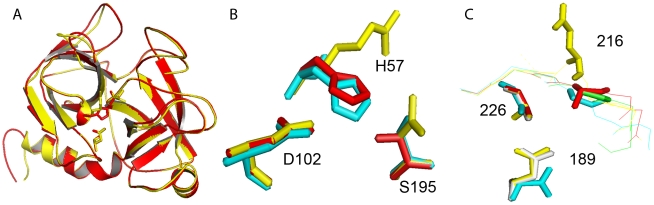
Figure 4. Structural models of rhinocerases.
Structural models of rhinocerases 2 to 5 were created using the IntFOLD server [17] using the structure of rat trypsin (PDB code: 1co9) as a template. A. Schematic diagram showing the overall similarities in structure between rhinocerase 2 (yellow) and rhinocerase 4 (red). The side chain atom positions for the catalytic triad residues are included. B. Detailed view of the amino acids corresponding to the catalytic triad residues in rhinocerase 2 (yellow), rhinocerase 4 (red) and chymotrypsin (PDB code: 1yph; cyan). Rhinocerase 2 has substitutions for the serine and histidine residues. C. Detailed view of the main constituents of the S1 specificity pocket in rhinocerase 2 (yellow), rhinocerase 4 (red), chymotrypsin (cyan) and trypsin (pdb code: 1co9; green). In chymotrypsin these residues are: S189 at the base of the specificity pocket, with G216 and G226 at the sides. In trypsin D189 is at the base of the pocket, with G216 and G226. All images were generated using PyMOL.