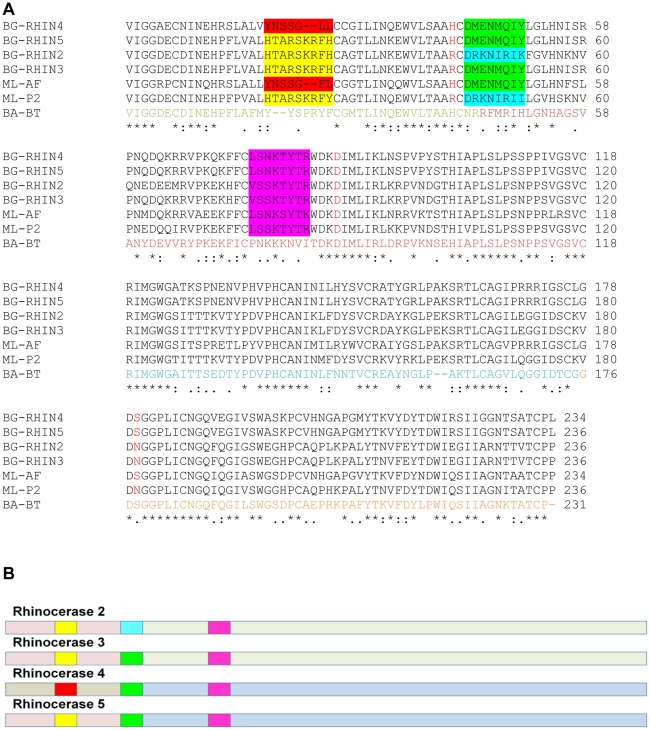
Figure 5. Evolution of rhinocerase enzymes.
A. Amino acid sequence alignment of rhinocerases 2 to 5, M. lebetina α-fibrinogenase (ML-AF) and venom serine proteinase-like protein 2 (ML-P2), and Bothrops atrox batroxobin (BA-BT). The alignment was created using ClustalW. The amino acids in the batroxobin sequence are coloured according to the exons encoding them: amino acids encoded by exon 2 are coloured green, and residues encoded by exons 3 to 5 are coloured dark red, blue and orange respectively. The catalytic triad residues are coloured red in the B.s gabonica and M. lebetina sequences. Coloured shading is used to indicate the three surface segments within serine proteases identified by Doley et al. [26] as undergoing accelerated change (ASSET). Residues in the first surface segment are shaded yellow or red depending on sequence similarity; residues in the second segment are shaded green or turquoise and the residues in the third segment are shaded pink.B. Schematic diagram of rhinocerases 2 to 5 indicating the different regions described in the text. The light shading represents the regions of each sequence corresponding to exon 2 and exons 3 to 5. Rhinocerases 2, 3 and 5 have similar N-terminal regions corresponding to exon 2 (pink), while rhinocerase 4 has a different sequence in this region (grey). In the C-terminal regions, corresponding to exons 3 to 5, rhinocerases 2 and 3 are similar (pale green) and rhinocerase 4 and 5 are similar to each other (pale blue). The brighter shading represents the three surface segments as shown in A. The first surface segment is identical in rhinocerases 2, 3 and 5 (yellow), and different in rhinocerase 4 (red). The second segment is identical in rhinocerases 3, 4 and 5 (green) and different in rhinocerase 2 (turquoise), and the third segment is very similar in all 3 sequences (pink).