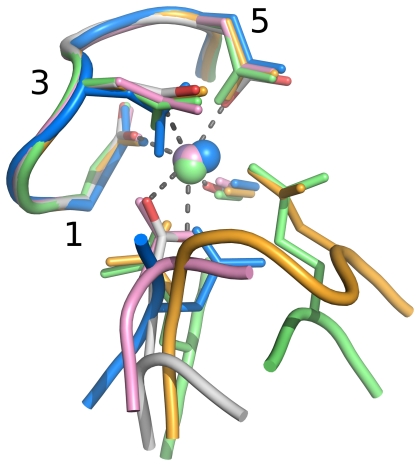
Figure 1. Comparison of Dx[DN]xDG calcium-binding motifs in calmodulin and the new structural contexts presented here.
The metal (sphere) is bound by the side chains of the Dx[DN]xDG motif (labelled 1, 3, 5) and the carbonyl group of the residue immediately following the motif. These, and the entire motif backbone, superimpose very well, while additional contributions to binding from later residues vary hugely in spacing and number (see text, Table 1 and Fig. 2). The representative calmodulin (PDB code 1exr) is coloured by atom type, with carbon white, oxygen red and bound calcium in purple. Other structures and their bound calcium ions are coloured uniformly with T. kodakaraensis subtilisin (PDB code 2z2x) in orange, endo-α-N-acetylgalactosaminidase (PDB code 2zxq) in pink, E. coli YgjK (PDB code 3c68) in green and the Porphyromonas adhesion domain (PDB code 3km5) in blue. Interactions of calmodulin with bound metal are shown as dotted lines.