Figure 2. Proposed model for the regulation of SMC specific genes during ESC/SMC differentiation and phenotypic switching.
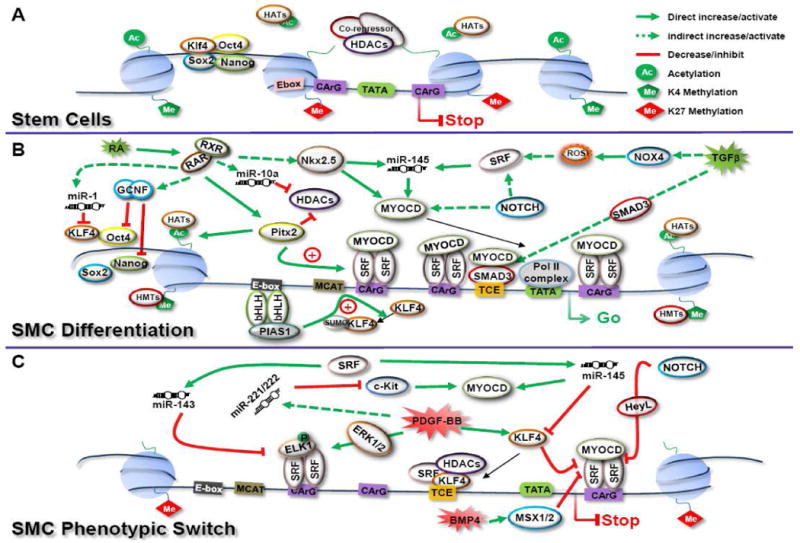
(A) In undifferentiated ESCs, Oct4, Sox2, Nanog and Klf4 form a core transcription complex maintaining the pluripotent network for stem cell self-renewal. Downstream genes associated with stem cell proliferation and pluripotency are actively transcribed marked with histone 3 lysine 4 methylation (H3K4me) and histone acetylation. Meanwhile, target genes co-occupied by the core transcription complex and encoding SMC specific genes are transcriptionally silent marked with H3K27me associated with the recruitment of HDACs and co-repressors at the regulatory regions of these SMC specific genes. As a result, the regulatory DNA sequence of these SMC specific genes are wrapped in compacted chromatin and blocked the access from MYOCD-SRF complex and subsequent activation. (B) SMC differentiation from ESCs has been shown to be mediated by multiple mechanisms. Extracellular stimuli (including retinoid acid and various growth factors) initiate the permanent shutdown of activation of the pluripotent core transcription complex on pluripotent genes and lead to downstream domino cascades. Ultimately, these serial programs result in dramatic chromatin modification in these regions, marked by histone acetylation and H3K4me and release the compacted chromatin containing the regulatory domain of SMC specific genes to expose the regulatory domains to diverse activator networks, including transcription factors, reactive oxygen species, miRNAs and nuclear receptors, etc. The transcription of most SMC specific genes is mainly activated by SRF binding to CArG boxes located within the regulatory region of SMC specific genes, enhanced by MYOCD. Additional elements further enhance transcription, such as bHLH transcription factors via PIAS-1 and TGFβ control element via SMAD3. (C) SMC phenotypic switching is initiated by various extracellular cues. During this process, transcription of SMC specific markers is downregulated and SMCs undergo the phenotypic transformation from contractile to proliferative/synthetic status. The regulatory networks involved into this change include histone modification that reprogram to deacetylation and H3K27me further closing active chromatin; phosphorylation-ELK1 by ERK1/2 and thereby blocking MYOCD interactions with SRF; KLF-4 and HeyL blocking SRF binding to CArG boxes; and microRNA interference operating post-trasncriptionally on critical regulators. Oct4, octamer-binding protein 4; Sox2, SRY-box containing gene 2; KLF-4, Kruppel like factor 4; HATs, histone acetyltransferases; HDACs, histone deacetylases; HMTs, histone methylation transferases; RXR, retinoid x receptor; RAR, retinoic acid receptor; GCNF, germ cell nuclear factor; miR, microRNA; Pitx2, paired-like homeodomain 2; Nkx2.5, NK2 transcription factor related, locus 5; MYOCD, myocardin; SRF, serum response factor; NOX4, NADPH oxidase 4; PIAS-1, protein inhibitor of activated STAT-1; Pol II, polymerase II; ELK1, ETS domain-containing protein 1; HeyL, hairy/enhancer-of-split related with YRPW motif-like; SMAD3, mothers against decapentaplegic homolog 3; Msx, muscle-segment homeobox like protein; TCE, TGFβ control element. bHLH, basic helix-loop-helix; RA, retinoid acid; TGFβ, transforming growth factor β; PDGF, platelet-derived growth factor; BMP4, bone morphogenetic protein 4; ROS, reactive oxygen species; Me, methylation; Ac, acetylation; SUMO, sumoylation.
