Abstract
Intestinal bacteria form a resident community that has co-evolved with the mammalian host. In addition to playing important roles in digestion and harvesting energy, commensal bacteria are crucial for the proper functioning of mucosal immune defenses. Most of these functions have been attributed to the presence of large numbers of “innocuous” resident bacteria that dilute or occupy niches for intestinal pathogens or induce innate immune responses that sequester bacteria in the lumen, thus quenching excessive activation of the mucosal immune system. However it has recently become obvious that commensal bacteria are not simply beneficial bystanders, but are important modulators of intestinal immune homeostasis and that the composition of the microbiota is a major factor in pre-determining the type and robustness of mucosal immune responses. Here we review specific examples of individual members of the microbiota that modify innate and adaptive immune responses, and we focus on potential mechanisms by which such species-specific signals are generated and transmitted to the host immune system.
Keywords: Commensal bacteria, intestinal microbiota, SFB, Th17, intestinal epithelium
Introduction
Commensal bacteria are indispensable for the normal function of the mammalian organism. It is well known that our ability to extract energy and nutrients from food depends on the presence of resident gut bacteria, the intestinal microbiota. In the course of evolution, this symbiotic relationship has allowed the host to overcome dietary restrictions and take advantage of otherwise unusable resources and habitats and has provided the microorganisms with a safe and nourishing environment. The co-evolution has resulted in almost complete co-dependence of the respective parties. Beyond digestion and metabolism, the microbiota also play vital roles in the protection of the host from potentially pathogenic microbes. In principle, such protection can be executed by activation of specific arms of the immune system, resulting in a reduced microbial burden, or elicitation of target cell responses, e.g. in epithelial cells, that confer host tolerance to microbes that would, otherwise, be pathogenic. One of the major puzzles in mucosal biology has been how the host tolerates the enormous intestinal bacterial burden and the potential immunogenicity of these organisms. Not much is currently known about host regulatory programs that contribute to tolerance towards commensal bacteria without reducing their numbers, although recent studies suggest that such programs do exist [1]. However, there has been substantial progress in understanding some of the ways used to keep pro-inflammatory immune mechanisms in check under normal physiological conditions. These involve establishment of immunological “ignorance” through physical sequestration of the bacteria in the lumen and induction of anti-inflammatory processes in the mucosal tissues.
It is now also appreciated that commensal bacteria have important functions in modulating the homeostasis of mucosal immune cell subsets, and specific examples have recently been described. These functions of commensals, albeit subtle at steady state, can have profound consequences during environmental challenge, and therefore affect the immunological fitness of the organism. They appear to be mediated by unique bacterial products, suggesting that microbial components that elicit specific host immune functions have been evolutionarily selected. Here we review these functions of the microbiota, and focus on potential mechanisms for maintenance of homeostasis by specific members of this community.
Commensal bacteria promote epithelial barrier function and evoke immune system-mediated protection of the mucosa
Sequestration in the lumen has been postulated to keep the systemic immune system generally unresponsive to commensal bacteria by preventing the two components from coming in contact under normal circumstances [2]. This state is known as immunological ignorance and is distinct from immunological tolerance, which involves the directed deletion or inactivation of cells with antigen-specific immune receptors [3,4]. At least some of the mechanisms securing the physical separation are induced by the bacteria themselves, possibly as an evolutionary adaptation to preserve mutualism. Signals from intestinal bacteria may promote integrity of the epithelial barrier and have been shown to regulate tight junctions and protect intestinal epithelial cells (IECs) from injury by controlling the rate of IEC proliferation and inducing cytoprotective proteins [5–9]. Commensal bacteria also induce the production and export in the intestinal lumen of secretory IgA (SIgA) and anti-microbial peptides (AMPs), such as RegIIIγ and alpha-defensins [10–13]. Microbial colonization stimulates the secretion of mucins from intestinal goblet cells. Mucins may participate directly in limiting intestinal infections by adhering to pathogens [14,15]. However, their main function is to form the backbone of the mucus covering the epithelial surface. The colonic mucus consists of two layers and although both layers contain MUC2 as a main structural component, the inner layer, which is firmly attached to the IECs is more viscous and difficult for bacteria to penetrate [16•]. The inner layer also serves to entrap most of the secreted anti-microbial peptides and SIgA, with the former being almost undetectable in the lumen [17]. As a result, the inner mucus layer is nearly free of bacteria [16•], preventing the bulk of the commensals from directly contacting the epithelial surface and the immune system. The importance of this sequestration is evident from studies in Muc2-deficient mice that develop spontaneous intestinal inflammation and increased susceptibility to intestinal infections due to an increased epithelial access of intestinal bacteria [18,19]. The mucus may also provide a medium in which bacterial-derived metabolites with important signaling functions are secreted and concentrated. Thus the mucus layer may promote mutualism by keeping bacteria at bay and restricting overt immune stimulation, while at the same time facilitating host-commensal or commensal-commensal crosstalk through the diffusion of bacterial products.
In spite of the mucus barrier and the containment of bacterial growth by AMPs, commensal bacteria are detected by the mucosal immune system. Direct sampling is mediated by M cells in Peyer’s patches, which is important for generation of antigen-specific responses, such as SIgA production. Luminal sensing may also occur through the extension of lamina propria (LP) dendritic cell (DC) projections into the lumen [20], which requires the presence of intestinal microbiota acting through TLRs [21].
Signals from intestinal bacteria are also continuously transmitted to the epithelium and are involved in maintaining tissue and immune homeostasis [22,23]. Loss of TLR signaling leads to exacerbation of intestinal inflammation when the epithelium is breached [9], and MyD88 ablation in Paneth cells results in loss of RegIIIγ production and defects in epithelial barrier function [24]. Commensal or non-pathogenic bacteria, mostly through TLR-mediated signals, can also inhibit NF-κB activation in IECs by multiple mechanisms [25]. These include inhibition of the pathway activator IRAK1 [26,27] and inhibition of the degradation of the pathway inhibitor IκBα [28] by microbiota-derived products, or the diversion of nuclear NF-κB by the peroxisome proliferator-activated receptor-γ (PPARγ) [29]. All of these mechanisms facilitate mutualism by decreasing tissue-damaging immune responses to commensal bacteria at steady state. What then is the role and relative importance of individual microbiota components?
Individual commensal bacteria modulate immune homeostasis through distinct mechanisms
It has been appreciated for a long time that beneficial bacteria exist, as exemplified by numerous studies on probiotics. Probiotics are bacterial species that exert beneficial health effects upon intestinal colonization. The model probiotics, Lactobacillus and Bifidobacterium spp, can inhibit the growth of intestinal pathogens by producing bactericidal compounds or metabolites that lower intestinal pH [30]. Other mechanisms such as induction of anti-inflammatory or protective cytokines have also been proposed, although the exact effects and mode of action of individual probiotics are likely to be quite different and are largely unknown. At the other end of the spectrum, the outgrowth, or loss, of certain components of the microbiota correlates with intestinal disease in both mouse and human [31–33]. In animal models, pathogenic changes in the composition of microbiota have been found upon breakdown of host immune homeostasis mechanisms [34], and in humans there can be similar dysbiosis following treatment with antibiotics, particularly in immunocompromised individuals [35]. Moreover, it has been possible to identify individual commensal components associated with or responsible for these effects, such as the loss of Faecalibacterium prauznitsii-mediated protective effects in inflammatory bowel disease patients [36•], or the expansion of pro-collitogenic Klebsiella pneumoniae and Proteus mirabilis in the Tbet−/−Rag−/− mouse spontaneous colitis model [37••].
Several studies have shown that the composition of the microbiota can influence obesity and energy balance [38] [39], and, more recently, effects of the microbiota on immune homeostasis have also been demonstrated. The first such example was the finding that mono-colonization of germ-free mice with a human commensal, Bacteroides fragilis, reversed the Th1/Th2 balance by inducing systemic Th1 responses [40]. This function was mediated by polysaccharide-A (PSA), a unique surface polysaccharide of B. fragilis. This bacterium was subsequently shown to induce anti-inflammatory responses and to alleviate intestinal inflammation through a process dependent on the cytokine IL-10, which may be produced by newly-induced regulatory T cells (Treg) [41••,42]. Injection or feeding mice with PSA was sufficient to replicate the B. fragilis immune effects. Thus, a resident intestinal bacterial species, through the production of a unique product, can affect systemic T cell homeostasis and mucosal immune responses.
Because B. fragilis is a human rather than mouse commensal, its effects in the mouse may or may not reflect evolutionarily selected commensal functions. However, there are now examples of mouse commensal microbes that can modulate the homeostasis of murine intestinal mucosal immune cell subsets. The first such example is the segmented filamentous bacteria (SFB), which were found to regulate the abundance of lamina propria Th17 cells [43••,44••]. SFB embed into the membranes of ileal epithelial cells, and are likely to initiate signals in these cells by way of this association. Although SFB were also reported to influence the abundance of other T cell subsets [44••], possibly due to differences in strains used, they have been specifically associated with Th17 cell numbers in several studies [43••,45•,46•]. Colonization of germ-free mice with a number of other defined commensal species or diverse microbiota lacking SFB did not induce Th17 cell differentiation [43••,47•]. Moreover, colonies of conventionally raised mice that possess diverse microbiota, but lack both Th17 cells and SFB, have been identified [47•]. The presence of SFB and Th17 cells in the context of normal microbiota was shown to modulate the nature of preexisting immune responses. Thus, SFB colonization influenced the proportion of systemic Th17 cells and exacerbated Th17 cell-mediated disease in mice with genetic predisposition to autoimmune arthritis or with induced experimental autoimmune encephalomyelitis (EAE) [46•,48]. At the same time, SFB colonization, possibly through the induction of IL-17 and IL-22, enhanced mucosal protection against an enteropathogenic bacterium, Citrobacter rodentium [43••]. Thus, although SFB does not induce a profound pro-inflammatory immune response, it affects the intestinal effector T cell balance, which in turn has significant consequences for the outcome of diverse immune challenges. This suggests that SFB may have been evolutionarily acquired for their effect on the host’s immunological fitness, and may contribute to the composition of the microbiota by restraining growth of potentially pathogenic microbes. SFB have been described in numerous vertebrate species, including mouse, rat, chicken, pig, and trout, but have not been described in human [43••]. Until now, only the 16S rRNA sequence of SFB has been available, and closely related 16S rRNA sequences have not been found in metagenomic studies of human microbiota, which may be due to limited sampling of humans. Alternatively, unique immunomodulatory mechanisms encoded by SFB may be conserved in related bacteria that colonize humans. Annotation of the full SFB genomic sequence may hence help identify such human commensals by exploring conservation of functional genetic modules in bacteria that colonize diverse mammalian species.
Regulatory T cells (Tregs) are another highly enriched T cell subset in the intestinal lamina propria, particularly in the large intestine. They are critical for the maintenance of intestinal homeostasis, and their numbers and phenotypes are also likely to be affected by signals from unique microbiota components. Although Foxp3+ Tregs are present and functional in the absence of microbiota [47•,49], gut bacteria may still modify the phenotype of Foxp3+ Tregs. In particular, inducible Tregs are generated from unpolarized CD4+ T cells in the intestine in response to production of TGF-β and, potentially, retinoic acid, by cells of the innate immune system. CD103+ lamina propria dendritic cells have been shown to encode the enzymatic machinery necessary to convert vitamin A to retinoic acid, and have been proposed to convey signals from the microbiota to direct Treg differentiation [50]. Natural Treg cells, generated in the thymus, are also found in gut mucosa, and may represent the bulk of Treg cells in the absence of microbiota or of specific components that induce Treg cell polarization. Commensal bacteria have been reported to induce not only Foxp3, but also IL-10 expression in the terminal ileum [44••], and production of this immunosuppressive cytokine may be critical for control of inflammation induced by potentially pathogenic microbes, e.g. Helicobacter in the colon [41••]. In our studies, the relative proportions of Foxp3+ Tregs were higher in the small intestine of germ-free animals compared to conventionally reared animals, and were reduced upon introduction of commensal bacteria [47•]. This agrees with reports that DNA derived from commensal bacteria suppresses Treg differentiation in a TLR9-dependent manner [51••]. Interestingly, the TLR9 effect is mediated by DNA from some, but not all, commensal bacteria [52]. Different rules appear to be in place in the colon, where commensal bacteria have a positive effect on Treg numbers. B. fragilis colonization has been reported to boost IL-10 production in Tregs in the colon in a PSA-dependent fashion [42], although the effect of PSA on Foxp3+ Treg proportions was marginal. A more comprehensive comparison of the effects of different commensal bacteria in a recent study demonstrated that a combination of spore-forming species of the genus Clostridium induced robust differentiation of inducible Foxp3+ and IL-10+ Tregs in the mouse colonic lamina propria [53••]. The Clostridium species belonged to clusters IV and XIVa. Interestingly, the aforementioned human commensal Faecalibacterium prauznitsii, which is decreased in Crohn’s disease patients and is associated with regulatory intestinal responses in humans, belongs to cluster IV and is phylogentically related to some of the mouse Treg inducing strains [36•]. The effect of the Clostridia was at least partially mediated by induction of TGF-β from IECs and boosting Clostridium numbers in neonatal mice increased resistance to colitis and suppressed systemic IgE responses [53••].
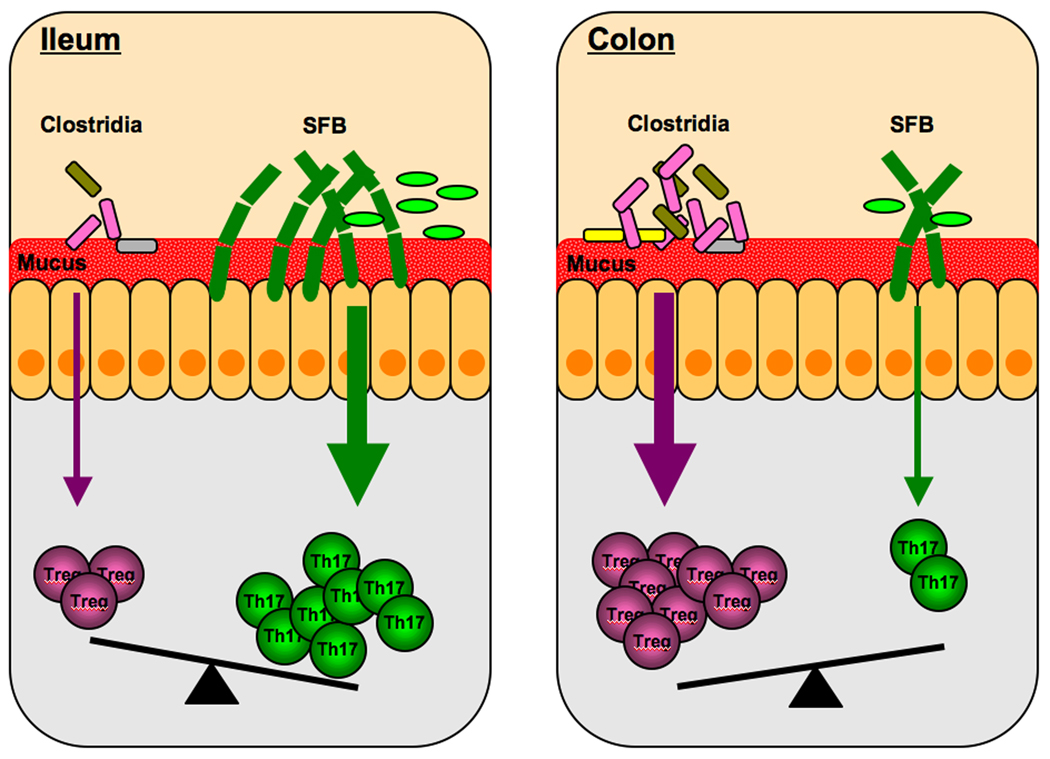
The studies described above mark the beginning of what will likely be numerous examples of immunomodulatory commensal species. The particular combination of these species in an individual or along the gastrointestinal tract would be expected to generate distinct immune environments in the host and in such way regulate immune responses. For example, the relative representations of SFB and cluster IV and XIVa Clostridia will control the balance between Th17 cells and inducible Treg and differences in the colonization abilities of these commensals may be responsible for the compartmentalization of mucosal immune responses, e.g. the prevalence of Th17 cells in the terminal ileum or Tregs in the colon (Figure 1).
Figure 1. Immunomodulatory commensal species control the Th17:Treg balance.
The balance between Th17 cells and Treg in the lamina propria is controlled by the relative representation of Th17 cell inducing species of the microbiota, such as SFB, and Treg inducing species, such as Clostridia from clusters IV and XIVa. In the small intestine, SFB, and possibly other unidentified Th17 cell-inducing commensal species, are overrepresented and Clostridia underrepresented, which leads to an abundance of Th17 cells in this location [43••]. Clostridia colonize preferentially cecum and colon, where they are responsible for the overwhelming representation of Treg [53••]. In this way, the composition of the microbiota directs the nature of the immune response in these two locations.
Mechanisms by which specific commensal bacteria affect immune homeostasis
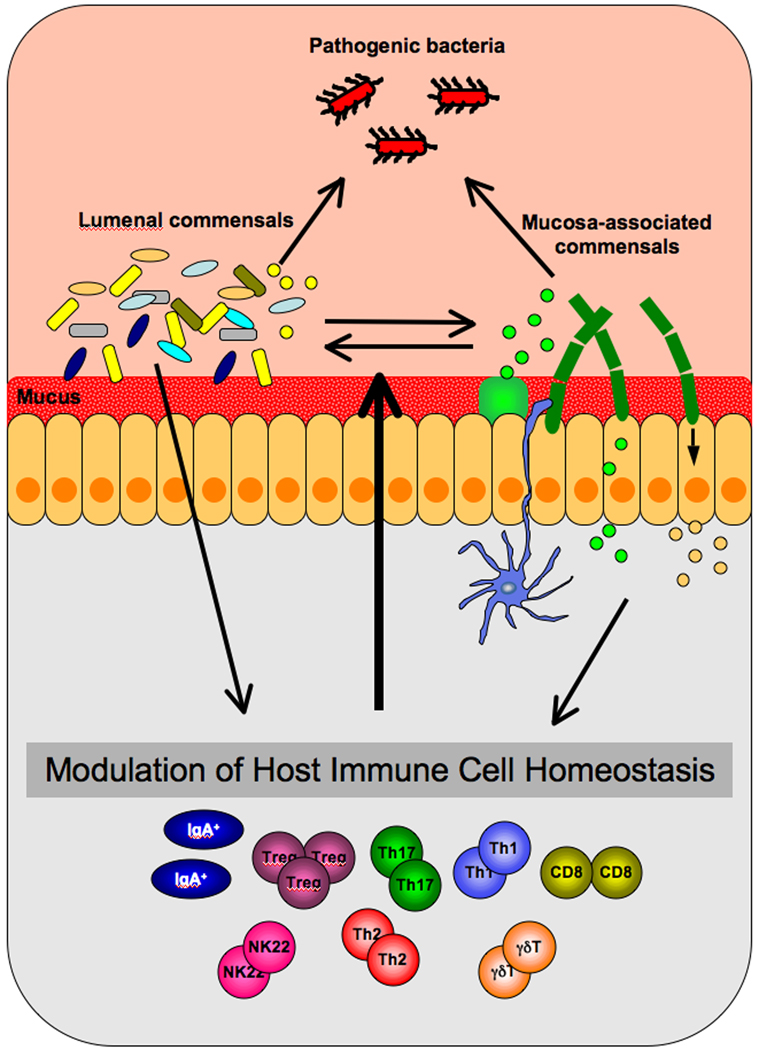
How may specific components of the microbiota exert their immunomodulatory effects? The interaction of SFB with the host organism presents the opportunity to study this problem in the context of induction of Th17 cells. A unique feature of SFB is their close association with IECs. SFB form long filaments, that attach to IECs in the terminal ileum [54]. Although the organization of the mucus may differ between the small and the large intestines, SFB must possess mechanisms for penetrating the mucus barrier. Such mechanisms have been described for other bacteria. For example Helicobacter pylori uses urease to lower mucus viscosity and Campilobacter jejuni and Salmonella spp have modified flagella for penetrating the mucus [55,56]. Once they penetrate the inner mucus layer, SFB surface molecules may engage specific IEC receptors to mediate anchoring and possibly also signaling, either through receptor ligation or through secretion of bacterial effector molecules that activate signaling pathways for production of cytokines. IEC cytokines have indeed been shown to be crucial for modulation of immune responses [13,57]. Mucosa-associated bacteria in general, and SFB in particular, could also be expected to be preferentially detected by interdigitating DCs and therefore may exert their immunomodulatory functions through DC signaling, activation, or antigen-presentation (Figure 2).
Figure 2. Modulation of immune homeostasis by signals from commensal bacteria.
Most commensals cannot penetrate the mucus barrier. However, certain members of the microbiota, such as segmented filamentous bacteria (SFB), can reach the epithelial surface and establish direct contact with the host tissue. SFB and other mucosa-associated bacteria may therefore engage receptors and downstream signaling pathways in intestinal epithelial cells (IECs) that can lead to the production of IEC cytokines. In addition, mucosa-associated bacteria may be readily detected by interdigitating dendritic cells. Both types of commensals may exercise their immunomodulatory effects through the secretion of specific metabolites. These metabolites would in turn engage IEC receptors or translocate to the lamina propria. They may also diffuse in the mucus layer and modify its local composition or affect signals from other commensal or pathogenic bacteria. Ultimately the integration of signals from multiple microbiota components can affect the homeostasis of effector immune cells in the lamina propria, which will dictate the nature of the host immune response during an environmental challenge, such as intestinal infection. Signals from immune cells in the lamina propria in turn affect microbiota composition and function in the lumen to help establish mutualism, immune balance and an individual’s level of protection.
Another mechanism by which SFB, and individual microbiota components in general, may induce specific immune functions is through the secretion or release of unique metabolites or metabolic enzymes. This mechanism allows for the generation of bacteria-specific substances that function as immune modulators. Because they are specific for particular bacterial taxa, from an evolutionary point of view, their integration into the host-commensal dialogue would be expected to help establish the bacterial taxon as a critical member of the microbiota. How this may be achieved can only be speculated on at this time. For example, SFB or SFB-induced cytokines, such as IL-17 or IL-22, may induce upregulation of SFB-binding receptors on IECs or the production of secreted products that may aid SFB colonization. Indeed, SFB induce both SIgA and AMP production [43••,58], which may help restrict competitive microbiota and provide a growth advantage for SFB. There are several ways by which bacteria-specific immunomodulatory factors may be generated: 1) the bacteria may secrete unique metabolic products; 2) the bacteria may release factors that modify host or other microbiota factors in the extracellular matrix to generate new molecules with unique signaling properties; 3) the bacteria may secrete enzymes that modify host or other microbes’ metabolic pathways and generate unique alternative byproducts or branches in a pathway that deplete original end products. The resulting unique metabolites can affect immune functions in different ways (Figure 2). They may bind to receptors on IECs and activate signaling pathways. They may penetrate the epithelium or be taken up by DCs, activating intracellular or cytosolic pathways. They may be deposited on extracellular matrix proteins, or on the surface of bacteria, changing their activity or their detection by immune cells. They may affect glycosylation patterns on surface receptors and hence modify their function. They may translocate to the circulation and affect systemic immunity. The possibilities go on and on.
Although studies examining the relative contribution of individual commensal species in vivo are at their onset, examples of bacterial taxon-specific metabolic products that affect mucosal and systemic immunity have been accumulating. PSA from Bacteroides fragilis, discussed above, is one example. Some probiotic and commensal bacteria secrete metabolites that can elicit responses in epithelial and other immune cells [59,60]. Peptidoglycan derived from certain commensal bacteria may enhance neutrophil function through Nod1 [61]. Commensal bacteria-derived ATP may enhance Th17 cell differentiation by activating P2X receptors on LP DCs [62•], and this may be a property of a distinct subset of intestinal bacteria. The ability of commensal bacteria to process nutrients leads to the generation of intermediates and metabolites that may have immunomodulatory functions. The glycan foraging ability of certain members of the microbiota is indispensable for the digestion of complex indigestible polysaccharides. Some commensal bacteria such as Bacteroides thetaiotaomicron possess an extraordinarily large collection of carbohydrate processing genes, which are absent from mammalian hosts [63]. One of the main byproducts of the activity of these members of the microbiota are short-chain fatty acids (SCFA) [64], which are important modulators of immune processes including chemotaxis, proliferation and cytokine production, and have anti-inflammatory effects on both colonic IECs and lymphocytes [65–67]. Commensal production of SCFA by certain beneficial bacterial species has also been shown to alleviate intestinal inflammation mediated by the expansion of the colitis-causing commensals Klebsiella pneumoniae and Proteus mirabilis in the absence of innate immune defenses [68] underscoring the importance of commensal bioproducts in this crosstalk. Dietary metabolites may also influence the commensal-immune dialogue by affecting the composition of the microbiota, which in turn affects immune homeostasis as demonstrated by the loss of Th17 cell-inducing bacteria in the absence of retinoic acid due to dietary vitamin A restriction [69].
The above examples support the idea that metabolic products derived from different members of the microbiota can serve as important modulators of immune homeostasis. The integration of such products into the crosstalk between bacteria and the immune system may represent an evolutionary adaptation of distinct commensal taxons that contributed to their being adopted as members of the host microbiota. At the same time, in the spirit of co-evolution of host and microbe, it has been proposed that the need to preserve commensal gut bacteria with unique metabolic benefits (e.g. complex polysaccharide utilization) may have led to corresponding adaptations of the host immune system and may represent a major driving force in the evolution of mucosal immunity [70,71].
Final thoughts
Most immune responses induced by commensals activate mechanisms that prevent bacteria from coming into direct contact with the immune cells of the host. These include AMP secretion, SIgA production, mucus generation, etc. These responses help establish mutualism by making the bacteria “invisible” to the immune system. Evasion strategies are regularly used by intestinal pathogens. However, certain intestinal pathogens have adopted a different strategy, making themselves more “evident” to the immune system by inducing inflammation. They have also developed mechanisms to counteract the anti-bacterial effects of inflammation, thus gaining a competitive advantage in the gut microenvironment [72]. Salmonella tymphimurium, for example, combines the use of microbiota-derived metabolic products and host-derived reactive oxygen species induced by the inflammation following infection to establish a growth advantage and outcompete commensal bacteria [73••]. We propose that some commensal bacteria have developed mechanisms to specifically activate the immune system and affect immune homeostasis. These mechanisms may skew the intestinal immune balance toward one or another effector T cell subset, for example. Thus, regulatory T cells may be induced to suppress pro-inflammatory responses and counteract the tactics of inflammation-dependent pathogens, such as Salmonella, while protective adaptive immune mechanisms, e.g. Th17 or Th1 cell differentiation, may be evoked to increase host fitness in fending off infectious organisms that try to evade inflammatory responses. The particular assortment of taxons in the individual’s microbiota may therefore pre-determine the nature and strength of the mucosal immune response during an environmental challenge. The benefits of each of the induced mechanisms would help preserve the corresponding microbial entities in the commensal repertoire of the host species. At the same time, the subtlety of the induced changes in immune homeostasis may allow for a diverse intraspecies representation of these microbial entities. This may explain why, despite the presence of a core microbiome, there is a vast diversity in the microbiota composition between individuals [74••,75••]. This model of “flexibility within constraints” posits that microbiota specificities are preserved within the species, yet individuals have diverse combinations of these specificities and therefore have different immune responses under the same environmental challenge. In combination with the genetic diversity of the host, the breadth of colonization with microbiota would thus guarantee adequate immune protection for the population as a whole. Consequently, perturbations of this evolutionarily established balance at the population level could result in changes in immunological fitness of the species. For example, the loss from the human population of individual members of the microbiota with immunomodulatory functions, due to the increase of modern practices such as widespread antibiotic usage, may be expected to decrease immunological fitness and can explain the rise of certain diseases, as has been postulated by the “disappearing microbiota” hypothesis [76].
An evolutionarily selected, balanced microbial community is present in the host intestine where it performs vital physiological functions and modifies immune homeostasis. The identification of the relative contributions of the members of this community or their gene products as well as their mechanisms of immune modulation will be invaluable in generating new strategies to regulate mucosal and systemic immune responses in health and disease.
ACKNOWLEDGMENTS
I.I.I. is supported by the National Institutes of Health and the Crohn’s and Colitis Foundation of America and D.R.L. by the Howard Hughes Medical Institute and the Helen and Martin Kimmel Center for Biology and Medicine.
References
- 1.Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 3.Konrad A, Cong Y, Duck W, Borlaza R, Elson CO. Tight mucosal compartmentation of the murine immune response to antigens of the enteric microbiota. Gastroenterology. 2006;130:2050–2059. doi: 10.1053/j.gastro.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 4.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 5.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 6.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, Bleich A, Bruder D, Franzke A, Rogler G, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One. 2007;2:e1308. doi: 10.1371/journal.pone.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Looijer-van Langen M, Madsen KL. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 9.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 12.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 13.He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Linden SK, Florin TH, McGuckin MA. Mucin dynamics in intestinal bacterial infection. PLoS One. 2008;3:e3952. doi: 10.1371/journal.pone.0003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linden SK, Sheng YH, Every AL, Miles KM, Skoog EC, Florin TH, Sutton P, McGuckin MA. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009;5:e1000617. doi: 10.1371/journal.ppat.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. • This paper describes the organization of the colonic mucus into a loose outer layer and an inner layer that is firmly attached to the epithelial cells. Commensal bacteia were detected only in the outer layer and did not directly contact the host tissues; however this sequestration was lost in MUC2-deficient mice.
- 17.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, Andersson M. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 18.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 19.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 21.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells JM, Rossi O, Meijerink M, van Baarlen P. Microbes and Health Sackler Colloquium: Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 24.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 26.Lotz M, Gutle D, Walther S, Menard S, Bogdan C, Hornef MW. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chassin C, Kocur M, Pott J, Duerr CU, Gutle D, Lotz M, Hornef MW. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 2010;8:358–368. doi: 10.1016/j.chom.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan ZQ, Jones DP, Neish AS. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. Embo J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 30.Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7:503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 32.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Dore J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 34.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 36. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. • This study, together with other reports, correlated the absence of the human gut commensal Faecalibacterium prauznitzii with disease recurrence in Crohn’s disease patients. The anti-inflammatory activity of the bacterium was demonstrated in vitro and in vivo in the TNBS mouse colitis model and was assigned to secreted metabolites.
- 37. Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. •• This elegant study tracked down and identified Klebsiella pneumoniae and Proteus mirabilis as colitogenic components of the microbiota that expand in the TRUC model of colitis. These strains could also elicit colitis in WT SPF, but not GF mice, demonstrating that their colitogenic activity depends on the presence of other microbiota members.
- 38.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 39.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 40.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 41. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. •• This study provided one of the first examples of how a symbiont-derived factor can affect mucosal immune responses and prevent intestinal inflammation. The human symbiont Bacteroides fragilis was shown to protect animals in several experimental colitis models. The beneficial activity was due to the unique B. fragilis polysaccharide (PSA) and required IL-10 production from CD4 T cells.
- 42.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. •• This study identified the first example of a specific immunomodulatory commensal-host interaction. Segmented filamentous bacteria (SFB), which are members of the mouse commensal microbiota, were found to specifically induce Th17 cells in the mucosal lamina propria independently of the presence of other commensal bacteria and therefore affect the immune balance of effector T cell subsets in the gut and influence mucosal immune responses.
- 44. Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. •• Together with the previous study, this paper identified SFB as an important immunomodulatory component of the mouse microbiota. In addition to Th17 cell induction, SFB was proposed to more generally activate the mucosal immune system. The study supports the idea of the existence of dominant evolutionarily conserved microbiota members with critical immune functions.
- 45. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2009 doi: 10.1038/ni.1825. • This study examined how Paneth cell α-defensins modulate the composition of the small intestinal microbiota, and, thus, indirectly regulate mucosal T cell differentiation. Increase in defensin expression was shown to eliminate mucosa-associated bacteria, such as SFB, which led to a specific decrease in Th17, but not Th1, cells in the lamina propria.
- 46. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. • This study provided evidence that specific changes in the composition of the intestinal microbiota can greatly affect extra-intestinal disease development. Presence of SFB in the mouse intestine was found to induce autoimmune arthritis through the induction of Th17 cells that collaborate with B lymphocytes to induce arthritogenic autoreactive antibody.
- 47. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. • This study demonstrated that the composition of the intestinal microbiota modulates the balance between Th17 cells and regulatory T cells in the lamina propria. Th17 cells were absent in germ-free mice, and were induced by commensal microbiota in mice from some vendors, but not others, indicating that one or more specific bacterial strains are responsible for this effect. SFB were later identified to be one of these strains.
- 48.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Microbes and Health Sackler Colloquium: Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chinen T, Volchkov PY, Chervonsky AV, Rudensky AY. A critical role for regulatory T cell-mediated control of inflammation in the absence of commensal microbiota. J Exp Med. 2010 doi: 10.1084/jem.20101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. •• This study showed that DNA from commensal bacteria modulate intestinal immune responses through TLR9. Commensal DNA was found to restrict Treg cell conversion and promote Th17 and Th1 cell responses. The study implicates TLR9 in a signaling pathway from commensal bacteria to Th17 cell induction.
- 52.Grainger JR, Hall JA, Bouladoux N, Oldenhove G, Belkaid Y. Microbe-dendritic cell dialog controls regulatory T-cell fate. Immunol Rev. 2010;234:305–316. doi: 10.1111/j.0105-2896.2009.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. doi: 10.1126/science.1198469. In Press. •• This study identified a combination of autochthonous Clostridium species belonging to clusters IV and XIVa in the mouse colon that are responsible for inducing regulatory T cells in the colonic lamina propria, providing another important example of immunomodulatory commensal species.
- 54.Blumershine RV, Savage DC. Filamentous microbes indigenous to the murine small bowel: a scanning electron microscopic study of their morphology and attachment to the epithelium. Microb Ecol. 1978:95–103. doi: 10.1007/BF02014280. [DOI] [PubMed] [Google Scholar]
- 55.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A. 2009;106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15:456–461. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 57.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menard S, Candalh C, Bambou JC, Terpend K, Cerf-Bensussan N, Heyman M. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53:821–828. doi: 10.1136/gut.2003.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. • This study showed the ATP produced by intestinal commensal bacteria can induce Th17 cell differentiation in the lamina propria through activation of P2X receptors on lamina propria DCs.
- 63.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 64.Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 65.Meijer K, de Vos P, Priebe MG. Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr Opin Clin Nutr Metab Care. 2010 doi: 10.1097/MCO.0b013e32833eebe5. [DOI] [PubMed] [Google Scholar]
- 66.Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13:2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cavaglieri CR, Nishiyama A, Fernandes LC, Curi R, Miles EA, Calder PC. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci. 2003;73:1683–1690. doi: 10.1016/s0024-3205(03)00490-9. [DOI] [PubMed] [Google Scholar]
- 68.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, Dubois A, Khlebnikov A, van Hylckama Vlieg JE, Punit S, Glickman JN, et al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cha HR, Chang SY, Chang JH, Kim JO, Yang JY, Kim CH, Kweon MN. Downregulation of Th17 cells in the small intestine by disruption of gut flora in the absence of retinoic acid. J Immunol. 2010;184:6799–6806. doi: 10.4049/jimmunol.0902944. [DOI] [PubMed] [Google Scholar]
- 70.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 71.Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol. 2009;7:367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- 72.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, Tsolis RM, Baumler AJ. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. •• This study identified a specific mechanism by which Salmonella utilizes gut inflammation and commensal bacteria to achieve colonization advantage. Salmonella established a growth advantage over commensal bacteria in the presence of tetrathionate, which was generated by the combination of host inflammation-induced reactive oxygen species and microbiota-derived thiosulphate.
- 74. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. •• This study examined the microbiota composition and the microbial gene content of a large cohort of individuals. Although microbiota species composition differed between individuals, there was an identifiable collection of shared microbial genes, labeled as the “core microbiome”.
- 75. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. •• This describes the largest current publicly available metagenomic sequence database, obtained from fecal microbiota of 124 individuals. The study identified a shared core microbiome of both bacterial taxons and microbial gene functions.
- 76.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]