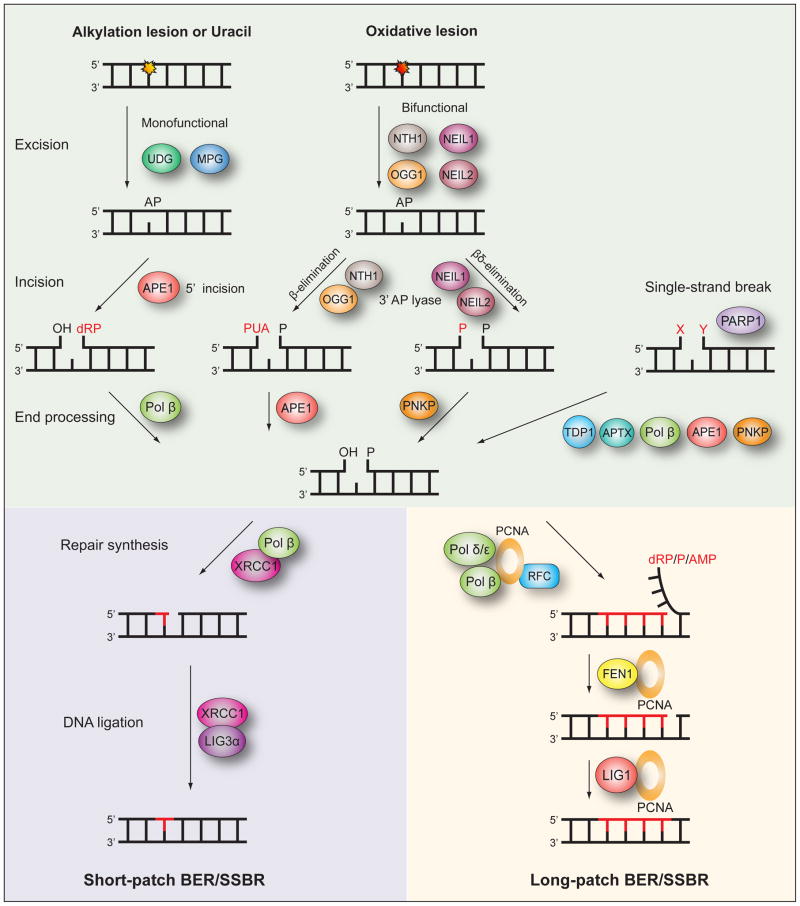
Fig. 3. The mammalian base excision repair and single-strand break repair pathways.
Base excision repair (BER) is initiated by removal of the modified base by either a monofunctional or bifunctional DNA glycosylase to leave an abasic site (AP). If excision is by either one of the monofunctional DNA glycosylases UDG or MPG, the following incision of the DNA backbone 5′ to the AP site is by APE1. Excision by one of the bifunctional DNA glycosylases NTH1, OGG1, NEIL1 or NEIL2 is followed by incision 3′ to the AP site via β- or βδ-elimination facilitated by the intrinsic 3′ AP lyase activity of these enzymes. The resulting single-strand break will contain either a 3′ or 5′ obstructive termini. End processing is then performed by Pol β, APE1 or PNKP depending on the specific nature of the terminus. Single-strand breaks do not only occur as intermediates of BER but also by other means and can contain simultaneous 3′ and 5′ obstructive termini. PARP1 recognizes these breaks and the end processing may utilize the additional factors TDP1 and APTX. When end processing has produced the necessary 3′-OH and 5′-P termini the following BER and single-strand break repair (SSBR) steps diverge into two subpathways, short-patch and long-patch. In short-patch BER/SSBR repair synthesis of the single nucleotide gap is by Pol β aided by the XRCC1 scaffold, and subsequent ligation by LIG3α finishes the repair. In long-patch BER/SSBR repair synthesis of the 2–13 nucleotide gap is by Pol β, and/or Pol δ/ε aided by PCNA and RFC. A resulting 5′ flap is removed by FEN1 and the the final ligation step is by LIG1.